2023-07-26 Hits(587)
Hybridoma cell antibody gene sequencing refers to the use of professional degenerate primer design and sequencing schemes, while optimizing economic and time costs, to provide customers with fast and accurate antibody variable region and full-length gene sequencing services.
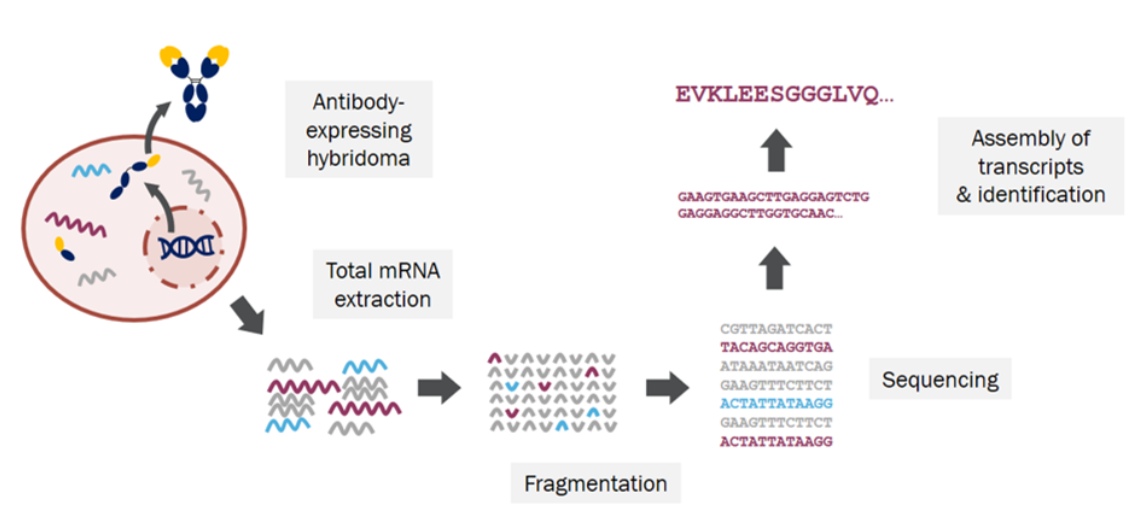
Figure 1: Hybridoma Sequencing
During the preservation process of hybridoma cells, there are cases of antibody gene loss, poor cell state affecting antibody production, and even cell death. To preserve excellent antibodies for a long time and stabilize production, antibody genes in hybridoma cells can be extracted and sequenced, ultimately obtaining nucleotide sequences, which are stored in DNA form for later exogenous expression and production.
The gene sequence of hybridoma monoclonal antibody obtained by hybridoma cell antibody sequencing is one of the prerequisites for publishing articles or patents and recombinant expression of engineered antibodies. It is also the basis for antibody humanization.
Standard cycle monoclonal antibody sequencing services: Antibody variable region VH and VL sequencing, antibody VH, VL and lead region sequencing, whole antibody VH, VL, lead region and constant region sequencing, completed within 3-4 weeks.
Rapid monoclonal antibody sequencing services: The antibody sequence rapidly recognized from the target cell line can obtain sequencing results within 10 days.
KMD Bioscience provides customers with hybridoma sequencing services, including: RNA extraction, DNA cloning, T vector construction, antibody gene sequencing, bioinformatics analysis and project report.
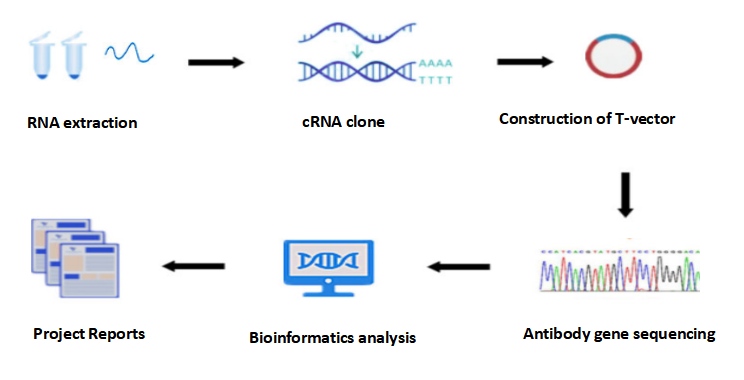
Figure 2: Hybridoma Sequencing process
Total RNA was extracted from hybridoma cells using TRIzol reagent according to the manufacturer's instructions.
A reverse transcriptase kit was used with additional RNA samples (hybridoma RNA samples from mouse antibodies or RNA samples from chimeric antibodies), primers, 10 mM deoxynucleotide triphosphate mixture (dNTPs), H2O, and 80U/ml RNASE inhibitor. Note: Due to its RNA content, the template was converted into oligonucleotides and stored at -80°C.
Perform the reverse transcription according to the following protocol: Keep all reactions on ice during the operation. For each RNA sample, set up three cDNA synthesis reactions: one for the kappa strand, one for the lambda strand, and one for the heavy strand. Ideally, each antibody amplifies only one light chain.

Figure 3: Protein sequence comparison of the 3H4 kappa and 3H4 lambda variable regions
5.2.1.1 In the PCR tube, prepare Mix 1: 2ml 50ng/ml RNA, 1ml 10mM reverse RT primers based on antibody chains (such as mIGKRT, mIGLRT, or mIGHGRT for mouse antibodies) and 1ml 10mM dNTP. For an RNA sample, three tubes of Mix1 are required, each tube containing different reverse primers.
5.2.1.2 In a 0.5 ml Eppendorf tube, prepare Mix 2:1.95ml H2O, 2ml 5x SMARTScribe buffer, 1ml 20 mM DTT, and 0.3m 100mM template-converting oligonucleotides. The volume provided for Mix2 is used for one cDNA synthesis reaction, scaled up as needed, and three times the volume of Mix 2 is prepared per hybridoma RNA sample. A master mix of Mix 2 was prepared for all reactions.
5.2.1.3 The RNA Protein secondary structure was denatured by incubating the tube containing Mix 1 at 72°C for 3 minutes in a thermal cycler.
5.2.1.4 During Mix 1 denaturation, the following substances were added to Mix 2: For each cDNA synthesis reaction 0.25ml 80U/ml RNAse inhibitors and 0.5ml 100U/ml SMARTScribe reverse transcriptase.
5.2.1.5 6ml of Mix 2 was added to each tube of denatured Mix 1.
5.2.1.6 In a thermal cycler, the combined mixture was incubated at 42°C for 60 min, followed by incubation at 70°C for 5 min to terminate the reaction. The reaction was maintained at 4°C. PCR amplification was performed immediately after reverse transcription. No cDNA purification step is required.
5.2.2.1 Set up PCR reactions for each cDNA synthesis: 10ml 5x PCR buffer, 1ml 10mM dNTPs, 3ml cDNA synthesized by RT reaction, 2.5ml 10mM universal forward primer ISPCR, 2.5ml 10mM reverse PCR primers on the antibody chain (for example mIGKPCR, mIGLPCR, or mIGHGPCR for mouse antibodies), 30.5ml of H2O, and 0.5ml 2U/ml Phusion polymerase (or other high fidelity polymerase).
5.2.2.2 Step-down/step-down PCR was performed according to the following thermocycler conditions: 98°C for 30 seconds; 10 cycles of 98°C for 15 seconds, 63°C to 57.5°C for 30 seconds (0.5°Cper cycle) and 72°C for 30 seconds; 15 cycles of 98°C for 15 seconds, 56°C for 30 seconds, and 72°C for 30 seconds; Then 72°C for 7 minutes and kept at 4°C.
5.2.2.3 5ml of each RT-PCR reaction was run on a 1% agarose gel in 90V TAE buffer. Amplified mouse antibody products appear between 550 and 600 base pairs. Amplified human antibody products appear between 750 and 850 base pairs. Quick Load Purple 2-Log DNA Ladder (NEB, N0550S) was used as a standard.
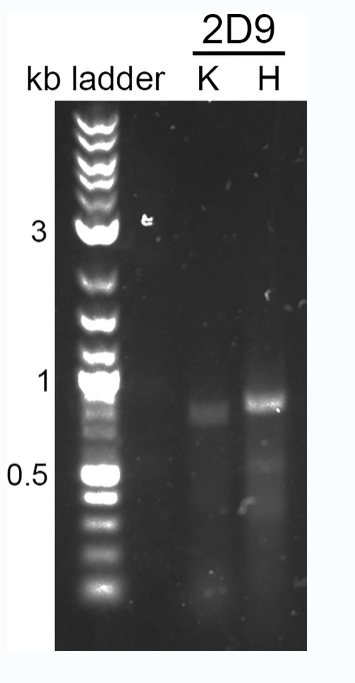
Figure 4: PCR amplification of the antibody variable region
Use the PCR cleaning and gel extraction kit of Macherey-Nagel to clean the RT-PCR reaction. According to the manual of the flat-end cloning kit, 2ml of each PCR-washed product was flat-ended cloned into the pCR-Blunt-II-TOPO vector. Next, 3ml of each TOPO cloning reaction was transformed into chemically competent E.coli. A total of 100ml of each transformation was spread onto LB plates containing 50mg/ml kanamycin and incubated overnight at 37°C.
After colonies were obtained, 5-10 colonies of each antibody chain were inoculated in 5ml LB/ kanamycin medium and shaken overnight at 37°C at 250rpm. These cultures were prepared in small quantities using Macherey-Nagel's small quantity preparation kit, and the resulting plasmid DNA was Sanger sequenced by Sequetech Corporation using M13 forward primers.
Sequencing data were analyzed using a custom Python program available on GitHub to remove non-functional DNA.
Delivery the antibody sequence reports. The acquisition of hybridoma cells is the basis for recombinant antibody production and antibody drug development. KMD Bioscience is able to accurately and skillfully sequence hybridoma cells from multiple species sources, including mouse hybridoma cells, rat hybridoma cells, rabbit hybridoma cells, and recombinant monoclonal antibodies of different species (obtained from the mAb gene constructed by phage technology). Through strict quality control, to ensure high accuracy.