2023-06-21 Hits(529)
Protein expression refers to a molecular biology technique that uses model organisms such as bacteria, yeast, animal cells or plant cells to express foreign gene proteins. Prokaryotic protein expression refers to the expression of foreign target genes in specific prokaryotes or cells by gene cloning technology, by constructing expression vectors and introducing them into expression strains.
Common protein expression systems include prokaryotic, yeast, insect, plant and mammalian expression systems, among which the most economical and affordable one is procaryotic protein expression system. The differences between the four common expression systems are as follows:
Table 1: Differences of Common Protein Expression Systems
|
Expression system |
Advantage |
Disadvantage |
|
Prokaryotic expression |
Economical and practical, with high expression. |
Glycosylation modification cannot be carried out to form inclusion bodies; Produce endotoxin. |
|
Yeast expression |
High expression, post-translation modification function, and large-scale expression. |
Lack of strong and strictly regulated promoters, low secretion efficiency. |
|
Mammalian expression |
The antigenicity and immunogenicity of the product are the closest to those of natural protein, and the glycosylation is accurate. |
Low yield. |
|
Insect expression |
The glycosylation form is low and the expression form is single. |
High cost and low expression. |
|
Plant expression |
Easy for large-scale culture |
Transgenic plants are expensive, the expression level is difficult to increase, and separation and purification are inconvenient. |
Protein expression level: prokaryote expression > yeast expression > mammalian expression. Insect system is close to prokaryote and yeast. Choosing a suitable protein expression system needs comprehensive consideration according to a series of factors such as your experimental cost and cycle. At present, prokaryote protein expression system is the most commonly used, and Escherichia coli and Bacillus subtilis are the common hosts of prokaryotic expression system. The former has high expression level, and endotoxin needs to be removed in later products. The latter has a relatively low protein yield, but does not produce endotoxin. The main difference is shown in the following figure:
|
project |
Escherichia coli expression host |
Bacillus subtilis expression host |
|
Advantages |
Wide application
simple operation
mass production
low cost. |
Low production cost
No endotoxin production
Can secrete expressed protein. |
|
disadvantages |
Poor secretion ability |
The output is relatively low. |
|
common hosts |
Rosetta(DE3)
Rosetta(DE3)pLysS
Rosetta2(DE3)pLysS
Origami2(DE3)
Rosetta-gami2(DE3)pLysS |
WB600
WB800N
Bacillus Subtilis 168 |
A complete protein expression system usually includes a host, foreign genes, vectors and auxiliary components.
The common host strains of protonucleoprotein are BL21(DE3) and Rosetta strains.
Because of the diversity of foreign genes, optimizing codons, improving the stability of foreign gene mRNA and optimizing vector design should be considered to express foreign genes efficiently.
Prokaryotic protein expression vector plasmids include replicon, promoter, selection marker, multiple cloning site (MCS) and fusion tag removal strategy. Common prokaryotic expression vectors are pET-28a(+), pGEX-4T-1 and pET SUMO. The prokaryotic expression plasmid vectors are as shown in the figure:
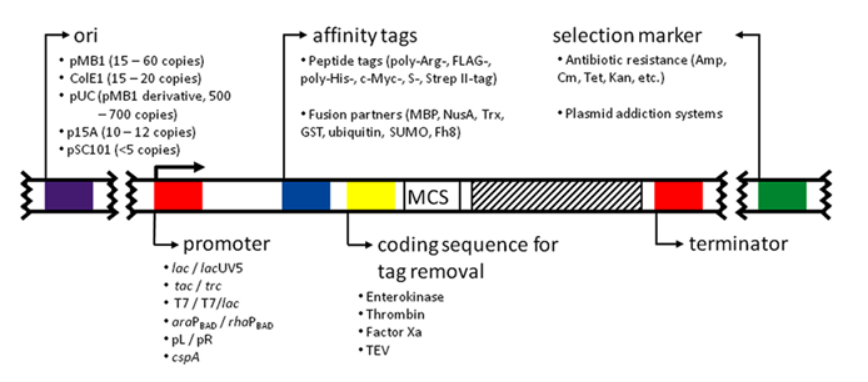
In the process of protein recombinant expression, the fusion expression of affinity tags has two functions: on the one hand, it is to make the protein purification process easier; On the other hand, it is to promote the solubility of some insoluble proteins. For the selection of fusion tag, please refer to the list of KMD Bioscience’s website.
Prokaryotic expression system has a very good application prospect. According to the relevant public information, the prokaryotic expression system can be used to produce recombinant proteins on a large scale, such as drugs, vaccines, antibodies, etc. (such as CMD-RP00002), thus providing strong support for the biomedical field. The prokaryotic expression system can be used to produce plant and animal growth regulators, antibacterial peptides, insect-resistant proteins, etc., thus improving crop yield and quality, reducing pesticide consumption and ensuring food safety. At the same time, prokaryotic expression system can also be used to produce industrial enzymes, biological dyes, etc., which has the advantages of high efficiency, low cost and environmental protection, and provides a new way for industrial production.
KMD Bioscience is committed to providing customers with high-quality prokaryote expression services, providing up to 400 prokaryote generation projects every year. KMD Bioscience has designed a set of effective expression vectors containing various fusion tags (His, GST, SUMO, FLAG, etc.), which can express recombinant proteins with molecular weight over 150kDa in a soluble way for customers. At the same time, we collected and established a variety of expression strains (including low-temperature induced expression strains, 10-16℃ ultra-low temperature induced expression proteins) to ensure the quality of each recombinant protein production.
The general experimental flow chart of prokaryotic protein expression is as follows:
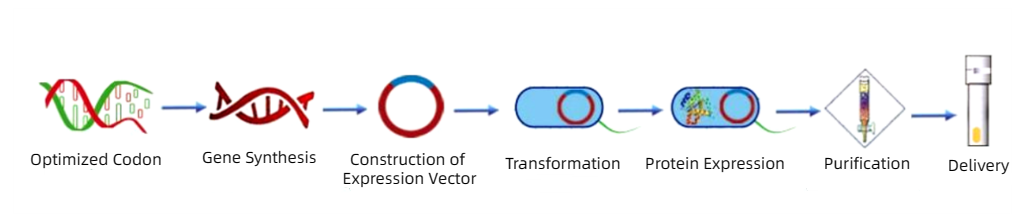
E. coli uses codons with different frequencies, including optimal codons and preferred codons, and generally chooses codons with a frequency greater than 20%. The optimization of gene sequence can improve the stability of secondary structure of mRNA, and avoid the delay/termination of translation, translation frame shift and amino acid mismatch due to insufficient number of tRNA libraries.
By PCR method, using the cloned plasmid containing the target gene as a template, a pair of primers were designed according to the gene sequence (different restriction sites were introduced into the upstream and downstream primers respectively), and the required gene fragments were obtained by PCR cycle.
Select appropriate vectors, common prokaryotic expression vectors are pET series, pGEX-4T-1, pET-SUMO, etc. The expression plasmid is double-digested with restriction endonuclease (restriction site with the same primer), the digested product is agarose electrophoresis, and the PCR product is recovered after double-digestion.
Prokaryotic proteins were produced by BL21(DE3) and Rosetta series strains. Plasmid vectors were transformed into RosseStta BL21 strain under the action of ligase. After screening with coated TB (containing 50ug/ml kana) plates, monoclonal clones were selected for activation culture.
When the OD600 of bacteria is 0.6-0.8, IPTG is induced to express at 37℃ for 4 hours, and the bacterial samples before and after induced expression are analyzed by SDS-PAGE. The results of induced expression are as follows:
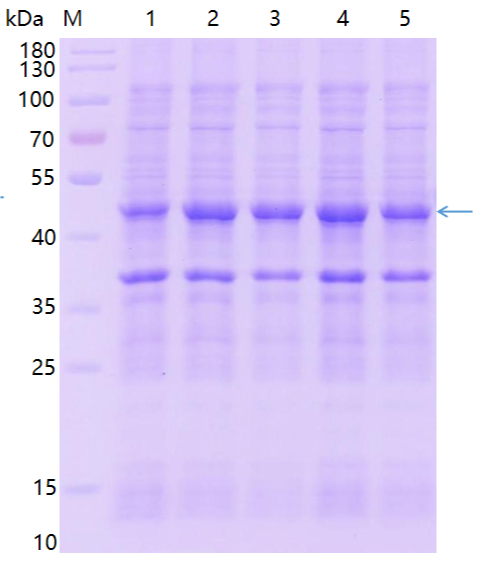
Take BL21 strain which can express the target protein, inoculate it into 1TB culture medium, add IPTG to induce it for 12h, centrifuge, collect bacterial sludge, and collect supernatant and precipitate after ultrasonic crushing. The induced expression results are amplified as shown in the following figure:
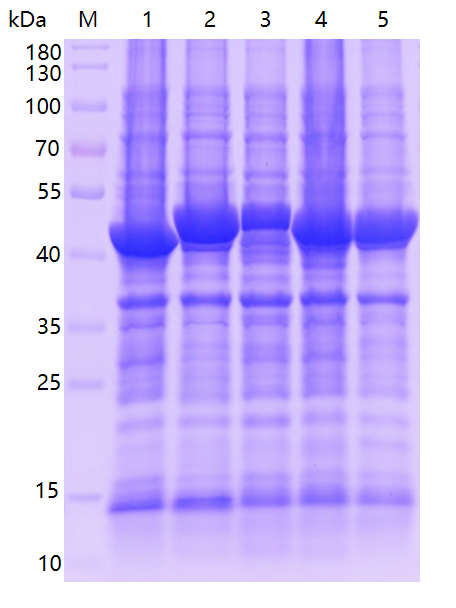
Purification of supernatant: Ni column affinity enrichment and purification on filtration membrane, SDS-PAGE analysis.
Purification of inclusion body (precipitate): buffer elution, Ni column affinity enrichment and purification, SDS-PAGE analysis, the purification results are as follows:
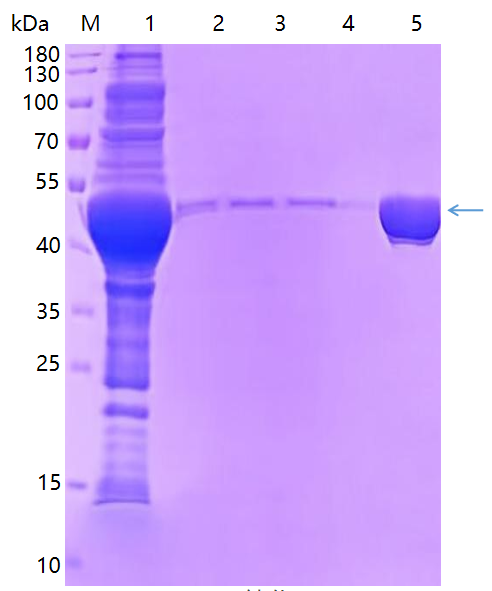
KMD Bioscience delivered complete experimental data report and determined concentration of protein, as well as SDS-PAGE result picture.
After years of development, KMD Bioscience has established a complete set of technological support for protein expression, protein fermentation, protein pilot and pilot scale. The recombinant expression and generation of protein in vitro usually involves the selection of expression host, gene synthesis, vector construction and other technical details. Customers only need to provide us with a protein sequence, CDS or protein name, and our experienced scientists can make a complete protein expression and purification plan for you in a short time.