2024-07-01 Hits(320)
Phage Display Technology
Phage display technology was born in 1985 when George Smith reported that foreign peptides could be displayed on the surface of filamentous bacteriophages [1]. Today, the phage display is a versatile tool for finding specific interactions between randomized library peptides/proteins on phage and target proteins, peptides, or other molecules [2].
By inserting foreign DNA into the genome of phages, fusion proteins are displayed on the surface of phages. The phage display library is a mixture of phages, each displaying different peptides/proteins enabling the selection of peptides/proteins with desired properties [3].
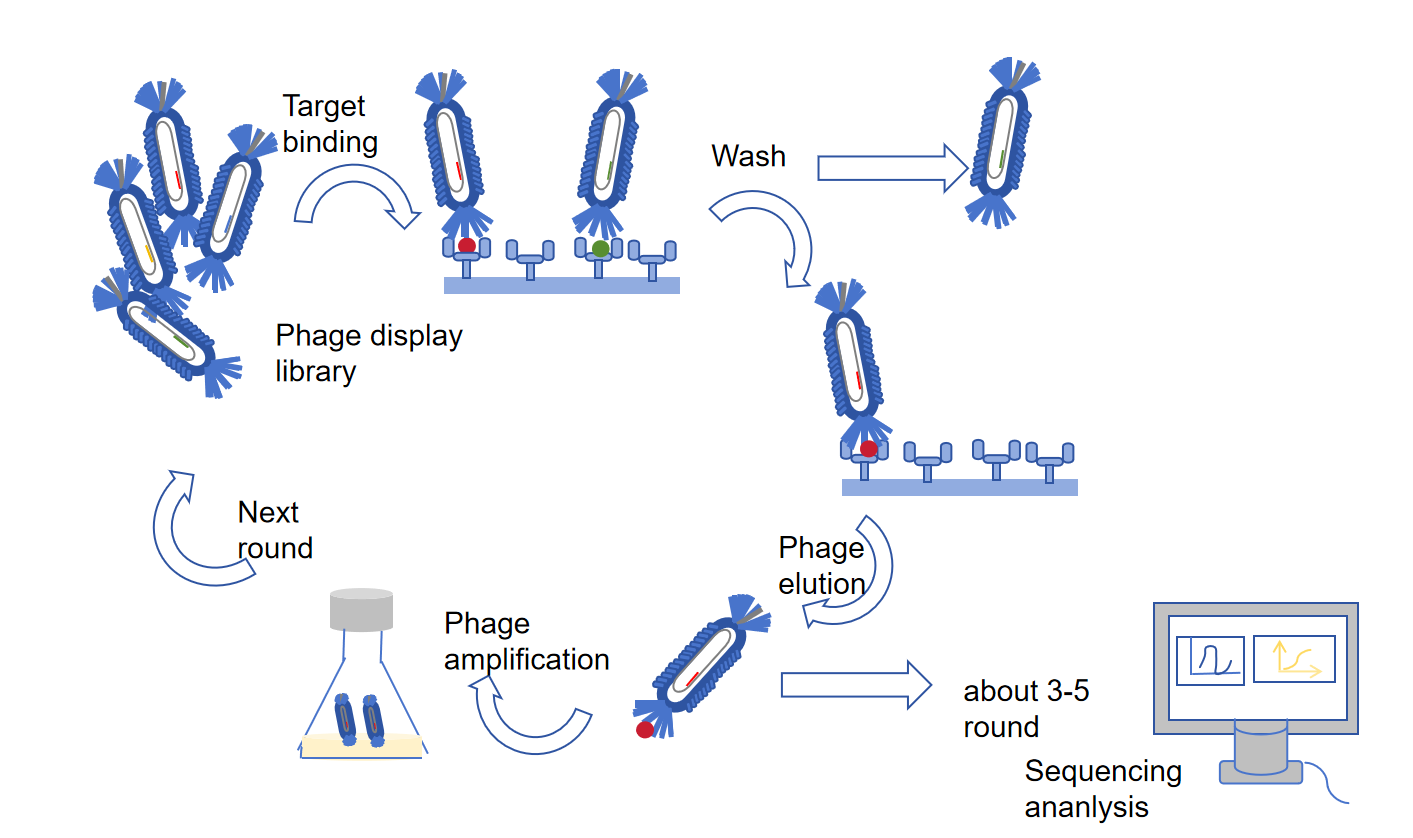
Figure 1 Typical procedure of phage library biopanning.
By using M13 phage to construct a phage display library, we can understand what M13 phage is.
The structure of M13 bacteriophage
The M13 bacteriophage is a non-lytic filamentous bacteriophage with a diameter of approximately 6 nm and a length of approximately 880 nm (Figure 1). 1A). Its clear genome is packaged in five capsid proteins (pIII, pVI, pVII, pVIII, pIX) and determines the length of the M13 bacteriophage [6]. Among the five capsid proteins, secondary capsid proteins pIII and pVI are capped at one end, while pVII and pIX are capped at the other end. Each secondary capsid protein has 3-5 copies. The main capsid protein pVIII consists of approximately 2700 copies, accounting for 98% of the total mass of M13 phage and containing 50 amino acids (AA). Each pVIII is composed of three structural domains: positively charged domains (40-50 AA) that interact with bacteriophage genomic DNA electrostatically, intermediate hydrophobic domains (21-39 AA), and N-terminal domains (1-20 AA). Through the electrostatic interaction between the phage genomic DNA and the positively charged domains of the pVIII protein, a total of 2700 pVIII protein copies spiral around the phage DNA, causing negative charges on the surface of M13.
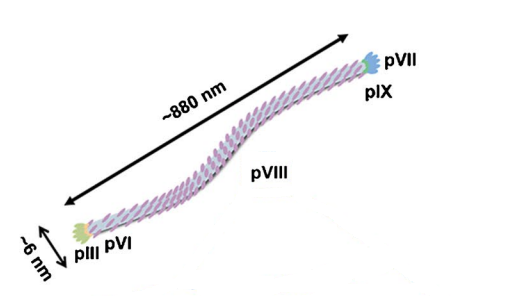
Figure 2 Illustration of the structure of M13 phage
The secondary capsid protein pIII is responsible for host cell recognition and infection. Although pIII only has 3-5 copies, it is one of the most widely used peptide display sites because its unconstrained protein structure allows for the insertion of over 100 exogenous peptides with AA. In addition to capsid proteins, there are several functional proteins responsible for M13 phage replication and assembly.
The most widely used model for constructing phage display libraries is the M13 bacteriophage. M13 bacteriophage is a non-lytic bacteriophage that displays proteins secreted through the periplasmic space of the Escherichia coli membrane and does not lyse Escherichia coli. The diameter is about 6nm and the length is about 880nm (Figure 2). Its genome is a circular single-stranded DNA molecule with a length of 6407bp, packaged in five capsid proteins [4].
Among the five capsid proteins, the secondary capsid proteins pIII and pVI, pVII and pIX are located at both ends, respectively. Each secondary capsid protein has 3-5 copies. The main capsid protein pVIII accounts for 98% of the total mass of M13 phage. Through the electrostatic interaction between the genomic DNA of bacteriophages and the positively charged domains of pVIII protein, a total of 2700 copies of pVIII protein spiral around bacteriophage DNA, causing negative charges on the surface of the M13 bacteriophage.
The secondary capsid protein pIII is responsible for host cell recognition and infection. Although pIII only has 3-5 copies, it is one of the most widely used capsid proteins for displaying peptides or proteins, as its protein structure allows for the insertion of over 100 amino acids [5]. Usually, pIII protein is used for low-price display (1-5 copies per phage), while pVIII protein is used for high-price display, with a maximum of 1000 copies per phage [6]. In addition to capsid proteins, there are several functional proteins responsible for M13 phage replication and assembly.
KMD Bioscience services
Phage display technology, peptide library screening services, and KMD Bioscience's antibody discovery platform are high-throughput screening methods. Through this technology platform, up to 10^13/ml of phage-displayed peptide or antibody clones can be constructed, achieving an average independent clone count of 10^9-10^10/ml. After multiple rounds of biological selection, up to 10^5 effective peptide or antibody sequences can be provided, providing a solid research foundation for subsequent enzyme substrate, a protein-ligand, and high-specific antibody searches. At the same time, with the affinity screening platform of KMD Bioscience, different peptides or antibodies can be obtained in a short period (2-3 days) for affinity sorting with target protein targets, and even precise affinity determination can be completed.
At present, based on the peptide library platform, KMD Bioscience can provide customers with random 7-peptide libraries, 12 peptide libraries, and cyclic 7-peptide library products for phage display, allowing customers to directly screen and obtain subsequent sequences. KMD Bioscience can also provide two types of phage libraries, M13 and T7, to construct reagent kits for customizing the library that customers want for subsequent screening.
Reference
[1] Fukunaga, K., & Taki, M. (2012). Practical tips for construction of custom peptide libraries and affinity selection by using commercially available phage display cloning systems. Journal of nucleic acids, 2012.
[2] Lubkowski, J., Hennecke, F., Plückthun, A., & Wlodawer, A. (1998). The structural basis of phage display is elucidated by the crystal structure of the N-terminal domains of g3p. Nature Structural Biology, 5(2), 140-147.
[3] Rondot, S., Koch, J., Breitling, F., & Dübel, S. (2001). A helper phage to improve single-chain antibody presentation in phage display. Nature Biotechnology, 19(1), 75-78.
[4] Smith, G. P. (1985). Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science, 228(4705), 1315-1317.
[5] Wang, R., Li, H. D., Cao, Y., Wang, Z. Y., Yang, T., & Wang, J. H. (2023). M13 phage: a versatile building block for a highly specific analysis platform. Analytical and Bioanalytical Chemistry, 1-18.
[6]Weiss, G. A., & Sidhu, S. S. (2000). Design and evolution of artificial M13 coat proteins. Journal of molecular biology, 300(1), 213-219.