2024-09-05 Hits(94)
Protein Sequencing
At present, amino acid sequencing is an important means of understanding protein structure and function. The N-terminus is the beginning of protein synthesis and an important structural and functional site for proteins and peptides. Due to the high specificity of protein N-terminal sequences, only a few amino acid residues are needed to identify most proteins through protein sequencing. N-terminal sequencing is mainly used to determine the beginning of protein amino acid sequences. The composition of the N-terminal sequence of a protein can affect its biological function. For example, the N-terminal sequence is related to the half-life and subcellular localization of the protein, and many post-translational modifications can occur at the N-terminus. Analyzing the N-terminal sequence of a protein through protein sequencing can reveal its biological function.
Protein N-terminal sequencing is widely used in biomedical research, disease diagnosis, protein breakage research, and drug development. Through N-terminal sequencing, the starting position of protein synthesis can be determined, which is beneficial for the study of protein synthesis mechanisms. The N-terminal sequence of proteins may change post-translational modifications, and these changes can be detected using N-terminal sequencing to identify whether the protein has been modified. N-terminal sequencing can also detect the presence and purity of specific proteins, which is beneficial for protein purification. By studying the N-terminal sequence of proteins, new drug targets can be discovered, providing new evidence for drug development. In addition, data from protein N-terminal sequencing can be used to predict protein structure, which is beneficial for scientific research such as protein fragmentation studies.
The principle of protein N-terminal sequencing
Protein N-terminal sequencing includes both non-mass spectrometry and mass spectrometry techniques. The principle of N-terminal sequencing is mainly based on the Edman chemical degradation method of non-mass spectrometry techniques, also known as Edman sequencing. Firstly, the N-terminus of the protein reacts with chemical reagents such as phenyl isothiocyanate (PITC) to form a stable phenylaminothiophthalein (PTC) derivative, known as the PTC peptide, which binds the N-terminal amino acid residues to specific chemical groups. Under alkaline conditions, PTC peptides undergo cyclization cleavage, producing an intermediate of thiazolidinone aniline (ATZ). The N-terminal amino acid residue is separated from the peptide chain, and its original linking sequence remains unchanged. The ATZ intermediate is subsequently converted into a stable and easily detectable compound, namely phenylisothiocyanine amino acid (PTH amino acid). The type of PTH amino acid can be identified through chromatographic or mass spectrometry analysis, and the amino acid sequence at the N-terminus of the protein can be determined through amino acid sequencing. The above process can remove an amino acid residue from the N-terminus of the protein in each cycle, and the next new free N-terminus will be exposed. Through continuous cyclic amino acid sequencing, the complete amino acid sequence of the protein N-terminus can be gradually determined. Although Edman sequencing has high accuracy, the proteins or peptides used for Edman sequencing must be of high purity, and Edman sequencing is only suitable for specific modifications of certain residues, without universal applicability. With the continuous advancement of mass spectrometry technology, various chemical modification techniques based on mass spectrometry are constantly developing and improving, deepening our understanding of the structure and function of proteins. At the same time, high-sensitivity, high-resolution, and high-throughput mass spectrometry technology also provides an important choice for protein N-terminal sequencing.
Sample requirements for protein N-terminal sequencing
The sample used for protein sequencing determines the quality of subsequent experimental results. The purity of the sample is generally greater than 90%, and some equipment may require a purity higher than 95%. The specific content of the sample may vary depending on the equipment, usually ranging from 50-100 pmol. The N-terminus of proteins used for sequencing must not be blocked or modified as it can hinder the Edman degradation process. Samples should try to avoid substances that may interfere with the sequencing process, such as proteases, amino acids, salts, acetaldehyde, etc. The sample types can be protein spots on PVDF membranes, protein freeze-dried powder, or naturally purified peptide samples, and different sample forms may require different processing methods.
When processing samples, the first step is to select appropriate extraction and purification methods based on the source of the sample. The purified protein needs to be dissolved in appropriate solvents such as distilled water, TFA aqueous solution, etc., and the solution should be adjusted to the appropriate concentration range. For samples on PVDF membranes, it is necessary to ensure that proteins are evenly distributed on the membrane, and glycine-containing buffer solutions that may interfere with sequencing should not be used during membrane transfer. During the process of handling samples, sterile operations must be performed to avoid contamination of the samples. The processed samples should be stored carefully to avoid high temperatures or strong light stimulation. Dry ice should be used for freezing protection during sample transportation to prevent degradation. When dissolving and diluting samples, avoid using salt solutions containing 1st and 2nd-grade amines, as well as solutions containing SDS, as they may interfere with sequencing results.
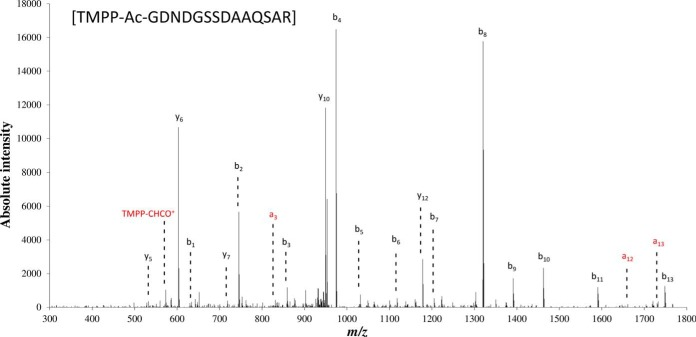
Fig. 1 N-terminal sequencing result chart
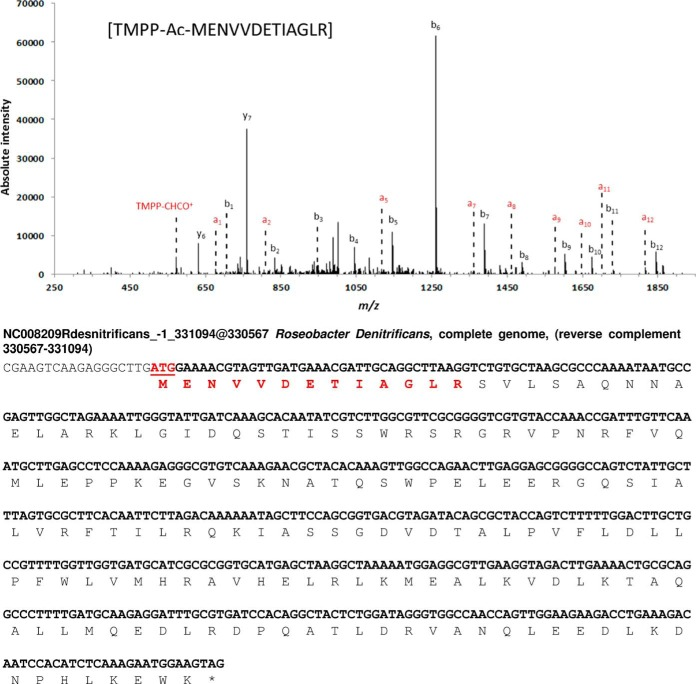
Fig. 2 Results of N-terminal sequencing MS/MS
KMD Bioscience has been committed to the research of recombinant protein expression and preparation for many years. In the process of analyzing the purified products of recombinant protein expression, it is necessary to confirm the N-terminal sequence of the protein. The N-terminal region is an important structural and functional site for proteins and peptides, and most proteins can be identified by a few amino acid residues at the N-terminus. We have experienced technicians, and high-quality sequencing instruments, and utilize existing mass spectrometry technology platforms to provide high-quality protein N-terminal sequencing services. Through the Edman degradation method, we can accurately determine protein N-terminal sequences of up to 67 amino acids. KMD Bioscience can perform N-terminal sequence validation of antibody drugs, identify naturally extracted and purified small molecule peptide sequences, and have a short experimental period of only 2-3 weeks.
Reference
[1] Bland C, Hartmann EM, Christie-Oleza JA, et al. N-Terminal-oriented proteogenomics of the marine bacterium roseobacter denitrificans Och114 using N-Succinimidyloxycarbonylmethyl)tris(2,4,6-trimethoxyphenyl)phosphonium bromide (TMPP) labeling and diagonal chromatography. Mol Cell Proteomics. 2014;13(5):1369-81.
[2] Speicher KD, Gorman N, Speicher DW. N-terminal sequence analysis of proteins and peptides. Curr Protoc Protein Sci. 2009;Chapter 11:11.10.1-11.10.31.
[3] Zhou C, Zhang Y, Qin P, et al. A method for rapidly confirming protein N-terminal sequences by matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun Mass Spectrom. 2006;20(19):2878-84.