2025-04-12 Hits(93)
Protein Sequencing
Principles and Methods of Protein Sequencing
Protein sequencing, the determination of amino acid sequence of proteins, is an important field in biochemistry and molecular biology. The structure of a protein determines its function, and the sequence of a protein is the basis of its structure. The goal of protein sequencing is to determine the sequence of amino acids in a protein molecule, that is, the sequence of proteins. Through sequencing technology, we can determine the amino acids in the protein one by one, thus revealing the sequence information of the protein. This information is critical to understanding the structure and function of proteins and provides a basis for further study and application of proteins[1].
The Principle of Protein Sequencing
1. Edman degradation method
This is a method of sequentially degrading proteins by chemical degradation starting at the N-terminal of the protein, removing one amino acid residue at a time, and identifying that amino acid. Phenylisothiocyanate (PITC) is used to react with the N-terminal amino acid of the protein to form a stable ring compound, which is then cut off from the protein under the action of the acid without affecting the rest of the amino acid sequence. This process can be repeated until the entire protein sequence is determined.
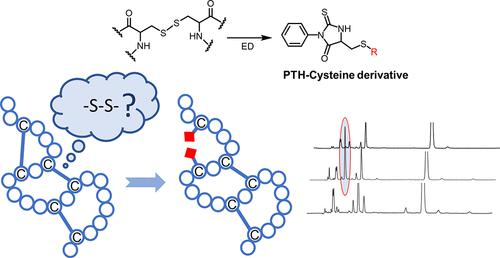
Figure 1:Edman degradation reveals unequivocal analysis of the disulfide connectivity in peptides and proteins[2]
2. Mass Spectrometry (MS)
The protein sample is ionized and then separated and detected according to the mass-charge ratio (m/z) of the ions in an electric or magnetic field. By analyzing the ion fragment patterns, the amino acid sequence of the protein can be deduced. Mass spectrometry can be used to determine the molecular weight of proteins and determine the de novo sequencing of proteins by Peptide Mass Fingerprinting or Tandem MS techniques. This method is fast and sensitive.
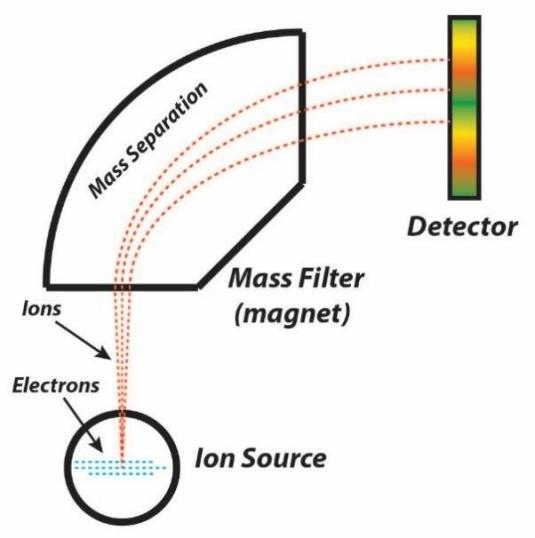
Figure 2:Schematic diagram of a mass spectrometer[3]
3. Amino Acid Analysis
Complete hydrolysis of a protein to a single amino acid by chemical or enzymatic reaction, and then analysis of amino acid composition and content using high performance liquid chromatography (HPLC) or other chromatographic techniques to infer the protein sequence indirectly.
4. DNA sequencing to infer protein sequences
Because proteins are encoded by genes, the corresponding protein sequence can be inferred by sequencing DNA or cDNA. This is usually done by first determining the gene sequence that codes for the protein and then inferring the amino acid sequence of the protein from the genetic code. DNA sequencing technology (such as Sanger sequencing, next generation sequencing, etc.) is used to determine the gene sequence, and then the amino acid sequence of the protein is translated through the genetic codon.
5. Protein Database Search
Use known protein sequence databases to infer the sequence of unknown proteins by comparing experimental data (such as mass spectrometry data).
Each method has its advantages and limitations, and usually in protein sequencing, the appropriate method is selected based on the specific needs of the study and the characteristics of the protein. For example, the Edman degradation method is suitable for sequencing small peptides, while mass spectrometry is more efficient for determining protein sequences in complex samples. In practice, these protein sequencing methods are sometimes used in combination to improve the accuracy and efficiency of sequencing.
Steps of Protein Sequencing
The steps of protein sequencing can vary depending on the method used. The following are the basic steps of the two main methods, Edman degradation and mass spectrometry:
1. Edman Degradation Process Steps
① Sample Preparation
Purify protein samples to ensure the purity and concentration of the samples are suitable for sequencing. If the protein is composed of multiple subunits, it needs to be dissociated into a single subunit first.
② Removal of N-terminal blocking amino acids (if present)
The N-terminal of some proteins may be closed (e.g. acetylation) and the blocking group needs to be removed by chemical or enzymatic methods.
③ Edman Degradation Cycle
Reaction of protein with phenyl isothiocyanate (PITC) to form n-terminal amino acid with PITC to form phenylthiocarbamide (PTC) derivative. Under the action of an acid, PTC derivatives are cut from the protein to form a ring of PTC-amino acids. This cyclic compound is isolated and identified by chromatography, thus determining the n-terminal amino acid. Repeat the above steps, cutting one amino acid at a time, to gradually determine the protein sequence.
④ Data Analysis
Stitch together the amino acid information from each cycle to get the complete protein sequence.
2. Mass Spectrometry Steps
① Sample Preparation
Purification of protein samples. It may be necessary to digest the protein into smaller peptide segments, usually using specific proteases (such as trypsin).
② Ionization
The peptide is ionized. Common ionization techniques include electrospray ionization (ESI) and matrix assisted laser desorption/ionization (MALDI).
③ Mass Spectrometry
The ionized peptide segment is introduced into the mass spectrometer for separation and detection according to the mass charge ratio (m/z). Molecular weight and fragment information of the peptide can be obtained by analyzing the mass spectrum of the peptide.
④ Sequence analysis
The amino acid sequence of the peptide segment is analyzed by database search or de novo sequencing using mass spectrometry data. If a database search is used, the experimental data needs to be compared with known sequences in the protein sequence database.
⑤ Splicing sequence
If a protein is digested into multiple peptide segments, the sequences of the different peptide segments need to be spliced together to obtain the complete protein sequence.
⑥ Data Analysis
Verify and confirm the sequence after splicing to ensure the accuracy of the sequence.
In practice, these steps may be adapted to specific experimental conditions and needs. In addition, with the development of technology, automated and high-throughput protein sequencing methods have gradually become popular, improving the efficiency and accuracy of sequencing.
Considerations for Protein Sequencing
Protein sequencing is a complex and accurate experimental technique, and the following aspects should be paid attention to when conducting protein sequencing.
1. Sample preparation:
① Purity: Ensures a high degree of purification of protein samples to avoid impurities interfering with sequencing results.
② Concentration: The concentration of the protein sample needs to meet the detection limit of the sequencing method, which usually requires micrograms or higher.
③ Integrity: Avoid degradation of proteins during extraction and purification and maintain their integrity.
④ Desalting: The removal of salt and other small molecular impurities from the sample to avoid interfering with the sequencing reaction.
2. Sequencing method selection:
① Suitability of methods: Appropriate sequencing methods were selected according to the properties of proteins (such as size, hydrophobicity, modification, etc.).
② Sensitivity: Consider the sensitivity of the sequencing method to ensure that low abundance proteins or peptides can be detected.
3. Experimental operation:
① Standardized operation: Follow standardized operating procedures to reduce experimental errors.
② Avoid contamination: Avoid cross-contamination during sample handling and sequencing.
③ Reaction conditions: Strict control of reaction conditions, such as temperature, pH, reaction time, etc., to ensure the specificity and efficiency of the reaction.
4. Data analysis:
① Software selection: Use appropriate software for data analysis and sequence analysis.
② Database comparison: If using a database search method, make sure to use an updated and comprehensive protein sequence database.
③ Verification of results: Verification of sequencing results, such as cross-validation by multiple methods or comparison using known standards.
5. Interpretation of results:
① Modification recognition: Pay attention to the identification of modified amino acids in proteins, such as phosphorylation, acetylation, etc., which may affect the sequencing results.
② Sequence coverage: Ensure that the sequencing results cover important regions of the protein, especially for large proteins or multi-subunit proteins.
Application of Protein Sequencing
Protein sequencing is very important in biomedical research and many applications. Here are some of the main applications:
1. Basic research
① Protein structure and function research: By determining the amino acid sequence of a protein, its secondary and tertiary structure can be predicted, and then its function can be inferred.
② Gene expression analysis: Protein sequencing can help verify the correspondence between gene expression products, namely proteins, and gene sequences.
③ Study of protein evolution and variation: By comparing protein sequences between different species or different individuals of the same species, it is possible to study the evolutionary relationship and variation of proteins.
2. Medical research
① Disease mechanism research: Many diseases are associated with abnormalities in specific proteins, and protein sequencing can help identify these abnormal proteins, thereby revealing disease mechanisms.
② Drug target discovery: By sequencing disease-related proteins, potential drug targets can be discovered to provide a basis for drug design.
③ Biomarker development: Protein sequencing facilitates the discovery and validation of biomarkers for disease diagnosis, prognostic judgment, and therapeutic efficacy monitoring.
3. Drug development
① Antibody engineering: When developing therapeutic antibodies, it is necessary to know precisely the amino acid sequence of the antibody in order to optimize its affinity and specificity.
② Protein drug design: Protein sequencing is the basis for designing protein-based drugs (such as enzyme replacement therapies, hormones, etc.).
③ Drug mechanism of action research: By sequencing the proteins after drug action, the mechanism of action and potential side effects of drugs can be studied.
With the continuous progress of sequencing technology and the reduction of costs, the application field of protein sequencing will continue to expand, providing more possibilities for scientific research and practical applications.
FAQ-Protein Sequencing
Q1: What are the advantages and disadvantages of Edman degradation?
A: Advantages: The sequencing is accurate and the N-terminal amino acid sequence can be determined.
Disadvantages: High sample purity is required, sequencing length is limited, and it is not suitable for modifying amino acids.
Q2: What are the advantages of mass spectrometry in protein sequencing?
A: High throughput, high sensitivity, complete protein and fragment determination, suitable for complex samples.
Q3: How to choose the right protein sequencing method?
A: According to the sample characteristics, sequencing purpose, equipment conditions and other factors selected. For example, for N-terminal sequencing, the Edman degradation method can be selected; For complex samples or full-length sequencing, mass spectrometry is more suitable.
Q4: How can protein sequencing results be verified?
A: The accuracy of sequencing results can be verified by database search, comparison of known sequences and bioinformatics analysis.
Q5: How to solve the problem of sample contamination in protein sequencing?
A: By optimizing extraction and purification steps, more rigorous sample handling methods such as chromatography techniques are used.
References
[1] Lavinder J J , Horton A P , Georgiou G ,et al.Next-generation sequencing and protein mass spectrometry for the comprehensive analysis of human cellular and serum antibody repertoires.[J].Current Opinion in Chemical Biology, 2015, 24:112-120.DOI:10.1016/j.cbpa.2014.11.007.
[2] Elsayed Y Y , Imhof D .Edman Degradation Reveals Unequivocal Analysis of the Disulfide Connectivity in Peptides and Proteins[J].[2025-02-08].
[3] Radauscher E J .Design, Fabrication, and Characterization of Carbon Nanotube Field Emission Devices for Advanced Applications[J]. 2016.DOI:10.13140/RG.2.2.16376.85767.