2024-07-26 Hits(528)
CRISPR/Cas9 Technology
CRISPR-Cas9 is the third generation gene editing technology developed after zinc finger endonuclease (ZFN) and transcription activator like effector nuclease (TALEN). As the most mainstream gene editing system today. CRISPR-Cas9 is one of the most efficient, simple, cost-effective, and easy-to-use gene editing technologies currently available. CRISPR-Cas9 first causes site-specific double strand breaks (DSBs) at specific locations in the genome, and then repairs the DSBs through non homologous end connections (NHEJ) or homologous recombination (HR), while completing gene editing.
Source of CRISPR Cas system
As one of the natural immune systems of prokaryotes, CRISPR/Cas is present in most bacteria and archaea. After a prokaryotic organism is invaded by a virus, due to its CRISPR/Cas system, a small segment of viral DNA is extracted and stored in a specific region of the prokaryotic organism's own genome, which is the CRISPR storage space. When the virus invades this organism again, the CRISPR/Cas system in prokaryotes recognizes the virus's DNA based on previously stored DNA fragments and cuts them off, allowing the expression of exogenous genes. This operation is similar to the principle of RNA interference (RNAi) in eukaryotes. Due to its precise targeting function, the CRISPR/Cas system has been developed as an efficient gene editing tool. There are currently three types of CRISPR/Cas systems: type I, type II, and type III. Among them, type II is relatively simple, consisting of Cas9 protein and guide RNA (gRNA) as the core, and is currently the most studied type.
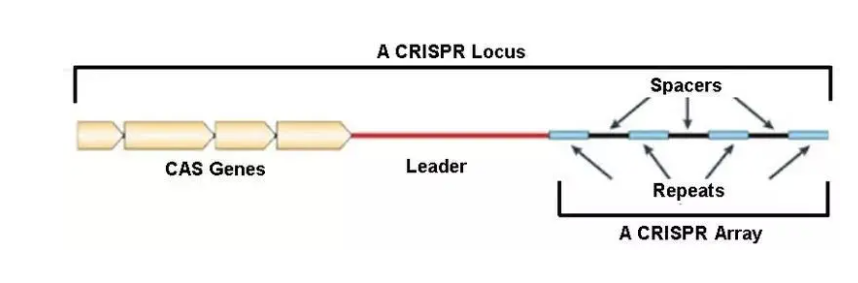
The composition of the CRISPR-Cas9 system
The CRISPR/Cas system derived from Streptococcus pyogenes is currently the most extensively studied system. Mainly including CRISPR loci and Cas genes (CRISPR associated genes).
CRISPR is composed of numerous short and conserved repeat regions and spacers. Repeats contain a palindrome sequence, forming a hairpin structure. However, Spacers are exogenous DNA sequences captured by bacteria, and each time an exogenous DNA sequence is obtained, it is integrated into the CRISPR sequence of the genome as a new spacer sequence. This is equivalent to the "blacklist" of the bacterial immune system. When the genetic material on these blacklists invades again, the CRISPR/Cas system will precisely strike and cut off the exogenous genes.
Meanwhile, the upstream leader region is referred to as the promoter of the CRISPR sequence. In addition, the Cas gene is located upstream of the CRISPR locus, and the proteins encoded by it can work together with CRISPR sequence regions. So far, various types of Cas genes such as Cas1-Cas10 have been discovered. In the actual working process, CRISPR sequences and Cas proteins cooperate with each other to perform defense functions in three steps. Cas9 protein performs DNA double strand cleavage function, while sgRNA (single guide RNA) has directing function.
CRISPR-Cas9 principle
The basic principle of CRISPR/Cas9 gene editing technology is that the Cas9 protein binds to artificially designed sgRNA to form an sgRNA-Cas9 protein complex, and under the guidance of sgRNA, exogenous DNA will be precisely cleaved by the Cas9 protein, causing double strand breaks (DSB).
After DSB induction, the organism itself has a response mechanism for DNA damage repair within the cell: non homologous end joining (NHEJ) and homologous directed repair (HDR). Without a template, NHEJ repair is used to connect the sequences at the upstream and downstream ends of the fracture or randomly insert or delete bases at the fracture site, causing gene inactivation. When a template is present, fragment insertion (Knock in) or site specific mutagenesis can be introduced at the cleavage site through homologous recombination.
The CRISPR/Cas9 system in organisms introduces mutations at specific sites through this method.
Advantages and limitations of CRISPR-Cas9
As the current preferred tool for gene editing, the CRISPR/Cas9 system also has many limitations. Firstly, there needs to be a relatively conservative PAM sequence (NGG) near the area to be edited. Secondly, the guide RNA needs to pair complementarily with the upstream sequence bases of PAM. To successfully recognize the target sequence, Cas9 protein must meet two conditions:
(1) base pairing between the 5 'end 20 nt of sgRNA and the target DNA;
(2) There is a suitable PAM sequence at the 3 'end of the target DNA.
The reason why CRISPR/Cas9 has overturned previous gene editing techniques is that it only requires changing the 5 'end sequence of sgRNA to reprogram Cas9's sequence specificity, making design and operation very convenient.