2022-06-09 Hits(745)
Monoclonal antibodies are the largest and fastest growing group of therapeutic proteins. More than 70 antibody drugs have received marketing approval for clinical use, and the market for antibody drugs surpassed $100 billion in 2017 . More than 500 therapeutic antibodies and their derivatives are currently in clinical trials, with a focus on cancer treatment and autoimmune diseases.
The first approved therapeutic antibodies were obtained through mouse hybridoma technology. However, murine antibodies elicit strong human anti-mouse antibody (HAMA) immune responses in patients because of their heterologous nature to humans. One solution to this problem is recombinant DNA technology. Using genetic engineering techniques, sequences of human antibodies are exchanged for partial sequences of mouse antibodies to obtain "chimeric" (mouse antibody constant region replaced by human antibody constant region) or "humanized" antibodies (only the mouse antigen binding region/complementary determining region CDR is transferred to the human variable region framework). These engineered antibodies dominated the first decade of therapeutic antibody approvals, creating a remarkable track record.
However, even the very highly humanized humanized antibodies still contain at least 1% ~ 5% heterologous components. As advances in fully humanized antibody technology have been made, fully humanized antibodies are emerging to dominate the development of new antibody drugs. Research on fully humanized antibodies began in the 1990s and has evolved into three representative technologies for preparing fully humanized antibodies: antibody library technology, transgenic animals, or transchromosomal animals. Transgenic animals, usually mice or rats, are experimentally introduced into the human immunoglobulin locus, while their own antibody genes are knocked out. Thus, after immunization, they use this sequence library to produce IgG from human germline sequences, from which monoclonal antibodies can be generated by classical hybridoma techniques. Antibody library technologies, such as the representative phage antibody display technology, can produce fully human human antibodies in vitro without immunization.
The preparation of fully human antibodies by phage antibody library technology usually starts with isolation of immunized or unimmunized B cells and amplification of all antibody VH and VL gene fragments therein by RT-PCR; the in vitro amplified VH and VL gene fragments are randomly cloned into the corresponding vectors and constructed into antibody combinatorial libraries in the form of Fab or ScFv; the antibody gene combinatorial libraries are then inserted into the phage-encoded membrane protein of gene III (g3) or gene VIII (g8) immediately downstream of the pilot series, so that the polypeptide expressed by the exogenous antibody gene can be displayed as a fusion protein at the N-terminal end of the phage shell protein pIII or pVIII. Each phage particle encodes and presents a different antibody (i.e., an "antibody library", Figure 1), which contains billions of individual clones. In these antibody libraries, genes encoding those antibodies that bind to antigen are screened for specificity and affinity by performing affinity enrichment-mild elution-phage amplification of the antigen in vitro, and then continuing to repeat the above enrichment screening process until a library of antibody phages with good specificity and affinity is obtained after several cycles, from which the genes for the V region of the fully human antibody are finally screened for good specificity and affinity (Figure 1).
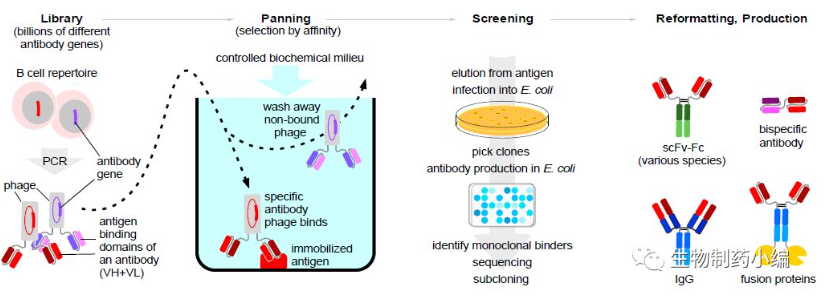
Figure 1. Phage antibody library technology
Classification of Phage Antibody Libraries
To date, researchers have created a large variety of antibody libraries that can be broadly classified into four categories: natural antibody libraries, immune libraries, semi-synthetic antibody libraries, and synthetic libraries. Natural antibody libraries are derived from hosts that have not been immunized with specific antigens. Theoretically, it is possible to screen natural antibody libraries for antibodies that specifically bind any antigen, but the lack of in vivo rearrangements and mutations in these antibody genes makes it relatively difficult to obtain high-affinity antibodies, and the screened antibodies require further affinity maturation modifications; and the screening background is high, with low abundance of antibodies to specific antigens.
Unlike natural antibody libraries, immune antibody libraries constructed by using host B cells derived from effective immunization against a specific antigen contain a large number of antibodies against that specific antigen, and the screening background is greatly reduced; and these antibody genes are often of satisfactory affinity after an in vivo affinity maturation process in the host. Usually using an immune antibody library with a library capacity of 1x10^6 clones, we can obtain antibodies with antibody affinity constants below 1x10^-9 mol/L higher affinity. Although suitable fully human antibodies can be easily obtained from a library of phage antibodies constructed by good antigen immunization, the preparation of fully human antibodies is often limited by the fact that humans cannot be immunized casually, which requires the creation of non-immune libraries with relatively large library capacity from which to screen the target fully human antibodies. Antibody libraries often require up to 1 x 10^12 clones to ensure that fully human antibodies with high affinity against specific antigenic epitopes are screened from them.
In addition, based on the framework region of natural antibody libraries and the CDR1 and CDR2 regions, semi-synthetic antibody libraries can be obtained by random synthetic amplification of the DNA sequence of the CDR3 region. Since CDR3 is the most important site for determining antibody specificity and has the greatest variation in structure and sequence diversity during individual B-cell genesis, it is most common to introduce random mutations in CDR3 while considering the variation in length. However, random mutations are introduced taking into account the relative conservatism of the CDR3 flankers and preserving certain residues that have an important role in antigen contact and structure formation. Synthetic libraries, on the other hand, are purely synthetic libraries designed and synthesized from antibody gene information.
Advantages and disadvantages of phage antibody library technology
The classical hybridoma technique requires a lot of time and effort to obtain high-affinity antibodies and requires subsequent humanization. Phage surface display technology can achieve short-term screening of higher affinity fully human antibody sequences without immunization and humanization modification steps, relying on enrichment screening in a fully controlled biochemical environment. This is unmatched by hybridoma technology. Phage display technology also allows access to some antibodies that are difficult to obtain by classical immunization, such as antibodies against very small or non-immunogenic or toxic substances, overcoming the barrier of difficulty in obtaining human-derived monoclonal antibodies by hybridoma technology.
However, it also has shortcomings, such as high requirements for library capacity. If the library capacity is small, it is difficult to obtain high-affinity antibodies; it is time-consuming and laborious to construct the library. The influence of a large number of unknown and unregulated factors on the library, in particular the potential limitations of diversity in IgM libraries (tendency towards monotypic expression of the V gene family) and the unknowability of the immune history of the B-cell donor, affect the capacity and quality of the library.
Classical screening strategy for phage antibody library technology
The classical screening method is based on incubation of a solid phase or liquid phase antigen with an antibody library and enrichment of specifically bound clones by several "adsorption-elution-amplification" cycles, which is simple, rapid and efficient. The following is a brief description of the classical screening methods.
1. Solid-phase antigen screening method
Antigens are directly encapsulated on the surface of solid phase media, such as enzyme labeling plates, immunotubes, and affinity columns, and antibody libraries are added for screening. Some antigens may be affected by their epitopes when wrapped on the solid phase media surface, and are screened by capturing the antigen with antibodies directed against other epitopes of the antigen (Figure 2).
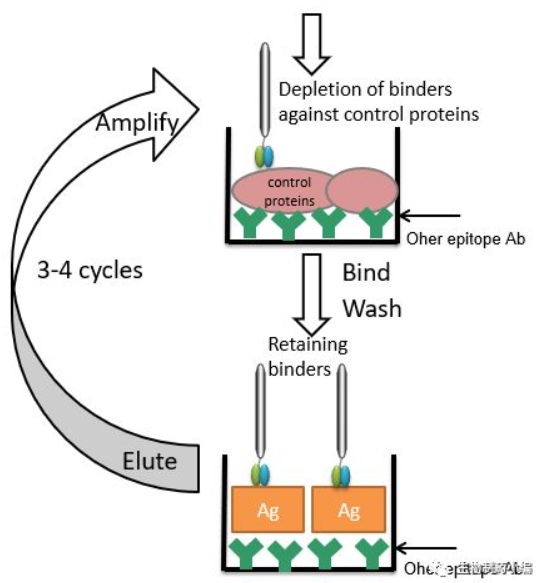
Figure 2. Solid-phase antigen screening method
2. Antigen liquid-phase screening method
Screening of phage antibodies in liquid phase using affinity magnetic beads after labeling biotin with antigen and with the help of magnetic field is a novel screening tool (Figure 3). It also solves the problem of disruption of the epitope of the solid-phase antigen.
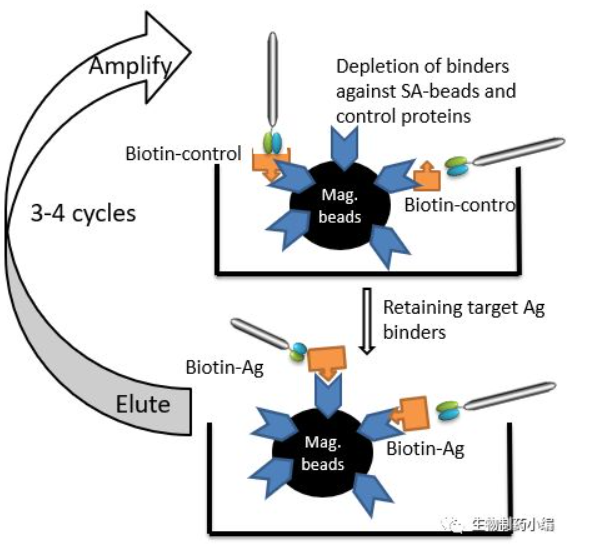
Figure 3. liquid phase antigen screening method
Concluding remarks
To date, six therapeutic antibodies generated by phage display have been approved for therapeutic use (Figure 4). Among them is the famous adalimumab Humira, which has been the global "drug king" for six consecutive years and achieved $18.427 billion in 2017 and is expected to exceed $20 billion in sales in 2018. As a powerful experimental technology in the post-genomic era, phage antibody library technology has the advantages of high efficiency, economy and speed, but there are still many aspects to improve in terms of affinity, library capacity and diversity. Therefore, it is imperative to develop ultra-high-capacity and high-affinity antibody libraries and to optimize affinity screening methods.
Reference
1. André Frenzel, Jonas Kügler. Designing Human Antibodies by Phage Display. Transfusion Medicine and Hemotherapy, 2017.
2.Mersmann M, Meier D, Mersmann J. Towards proteome scale antibody selections using phage display. New Biotechnol, 2010.
3.Bradbury ARM, Sidhu S, Dubel S. Beyond natural antibodies: the power of in vitro display technologies. Nat Biotechnol, 2011.
4.Hust M, Meyer T, Voedisch B, Rulker T, A human scFv antibody generation pipeline for proteome research. J Biotechnol, 2011.