2024-08-21 Hits(188)
Technology Service
The fluorescence in situ hybridization (FISH) technique is based on the known DNA probe, and RNA probe, and applying non-radioactive fluorescent substances, bacterial probe, and the target under test, the physical positioning of sequences of DNA on the chromosomes of technology, is one of the important experimental methods in molecular cytogenetics, has a good application prospect.
KMD Bioscience can provide models from probe design, sample processing, hybridization detection, and gene expression studies to data analysis and other one-stop FISH experiment design services, realize the standardization of the test procedure, and standardize, and improve the accuracy of experimental results, reduce the false positives and false negatives, ensure to serve customers with high quality of FSH.
The application of fluorescence in situ hybridization and detection range
Identify chromosomes
In the process of reproduction and evolution, whole-genome duplication and chromosome-doubling events often occur. Therefore, understanding chromosome ploidy and genomic karyotype analysis is essential to study the evolutionary history of species. We can use a specific DNA probe, bacterial probe, and RNA probe method to identify the same homologous chromosomes within a species and to determine the relationship between the chromosome ratio and FISH signal, to realize the different genome chromosome identification.
Karyotype analysis
Fluorescence in situ hybridization (FISH) technology uses non-radioactive fluorescent substances to label known nucleic acid probes and hybridize with the test target, to achieve accurate localization of DNA sequences on chromosomes. The length can be used as a chromosome and chromosome identification of important reference bases and is suitable for all kinds of different species of karyotype analysis.
Genetic relationship analysis
The degree of homology between target DNA and probe DNA can be directly reflected by the degree of chromosomal coverage or signal intensity of fluorescence in situ hybridization (FISH). So the technology can be directly applied to the analysis of genetic relationships between different species.
Identification of transgenes and heterologous chromosomes
In genetically modified samples or filters or allosomes additional to the genetic breeding materials research, exogenous gene fragment or exogenous chromosome identification is the foundation of scientific research. Fluorescence in situ hybridization (FISH) is an important means to physically localize DNA sequences on chromosomes and has become one of the key methods for identifying heterologous fragments within the genome. By fluorescence in situ hybridization technique can visually show gm integration sites, by the subsequent experiment.
The construction of a physical map and chromosomal location
Functional genomics research needs accurate genomic sequence information, so that map-based gene cloning, etc., to provide a real physical location on the chromosome. A genetic linkage map is assembled through genome sequencing and sequence when one of the important references is. By constructing genetic linkage maps based on recombination rates, we can learn that recombination events are not evenly distributed on chromosomes, which leads to a certain degree of difference between the distance of two loci on the genetic linkage map and their physical distance on the chromosome. Therefore, when carrying out genome sequence assembly and mapping cloning, we need to pay attention to the limitations of genetic linkage maps for guidance.
Probe type and preparation
The classification and preparation of fluorescent probe
According to different experimental requirements, the design of DNA probes will have a variety of types. These types include a single copy sequence of probe, the probe of the genome, a single copy of RNA probe, repetitive sequence, and oligonucleotide library probes. Probe in the repeated sequences with good repeatability high copy number, and signal strength is higher, is widely used in hybrid species.
These probes were prepared mainly by enzyme digestion amplification, PCR amplification, and chemical methods. Conventional repetitive sequence probes, genomic probes, and BAC library probes can be prepared using the above method.
Types and preparation methods of oligonucleotide probes
According to the design, the area can be divided into single-copy oligomeric nucleotide probes and repetitive sequence oligomeric probes. A repetitive sequence of oligomeric nucleotide probes can significantly shorten the hybrid length and can be used to detect repetitive sequence distribution in the chromosome and chromosome ploidy for rapid detection of species.
According to the probe, the design can be divided into paint Oligo - FISH and barcode Oligo - FISH two kinds.
The oligomeric nucleotide probe is also divided into single and double-stranded probes. Single probe preparation needs to be objective when the library after translation into RNA transcription, after marking fluorescent tags group, hybrid directly. Double-stranded probes need only PCR amplification with fluorescent marker primers.
The matters needing attention of all kinds of sample processing
If the test sample is materials (such as cell lines, chromosome preparation, etc.) in its application to the microscope slides, the need to use a mixture of methanol and acetic acid in a ratio of 3:1 fixed sample for 10 minutes. The fixed mixture should be ready in advance, and preparations after natural drying such as degeneration and hybridization experiments.
If the sample is paraffin section, FFPE tissue should be first cut into 5 μm thickness, placed on salinized or positively charged glass slides, and baked at 56 ℃ overnight, and then the material should be deparaffinized and pretreated.
Specific case analysis
rRNA of microbial detection and recognition, realization of in situ detection, identification, and choice of a specific group of bacteria and archaea. Certain bacteria and archaea groups showed different fluorescence.
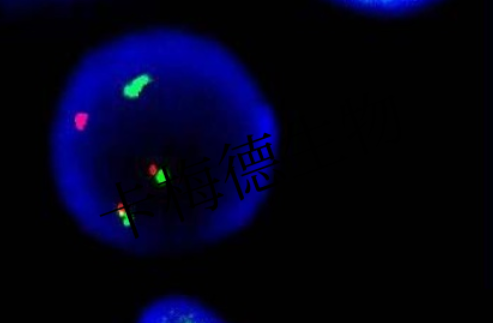
Figure 1. Microbial fluorescence in situ hybridization detection results
After years of scientific research efforts, KMD Bioscience has rich experience in FISH experiment designing, and the establishment of a comprehensive FISH technical services platform. We can provide one-stop FISH design services, including probe design, sample processing, hybridization detection, gene expression study, and data analysis. Through the standardization and standardization of test procedure, we improve the precision of the result of the experiment and reduce the incidence of false positives and false negatives, to ensure that provide customers with high-quality FISH technical services.
References
[1] Christine, E, Fuller, et al. Fluorescence in Situ Hybridization (FISH) in Diagnostic and Investigative Neuropathology[J]. Brain Pathology, 2021, 33(41):291-291.
[2] Pernthaler A, Pernthaler J, Amann R. Fluorescence in Situ Hybridization and Catalyzed Reporter Deposition for the Identification of Marine Bacteria[J]. Applied and Environmental Microbiology, 2020.
[3] Jiang J, Gill B S. Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research[J]. Genome, 2023, 49(9): 1057-1068.