2024-11-14 Hits(172)
In the field of yeast surface display library construction service, KMD Bioscience have rich experience and professional technical team to provide strong support for antibody drug discovery, disease diagnosis and biotherapeutics through innovative yeast display antibody discovery technology.
KMD Bioscience technical team consists of experienced engineers to ensure the professionalism and efficiency of our services. A comprehensive service process that provides a one-stop service experience from project consultation to final delivery. We have a strict quality control system to ensure the high standard and quality of our services. Moreover, we can provide customized services to meet different research and application needs.
KMD Bioscience can provide customers with high-quality yeast display services including VHH, scFv and other antibodies, and the yeast library capacity can reach 10^8, library diversity, insertion rate, positive rate can reach more than 90%, meeting the quality requirements of various customers for antibody yeast display libraries. At the same time, it can also provide one-stop technical services such as downstream antibody validation, antibody humanization, antibody affinity maturation, CAR-T/CAR-NK leader sequence design and cell killing verification for antibody yeast display.
Yeast Surface Display
Yeast surface display (YSD) is a widely used cell surface display technology. It is a technology that fuses the gene sequence of a foreign target protein with a specific carrier gene sequence into yeast cells, and then uses the protein transport mechanism in yeast cells to express the target protein and locate it on the surface of yeast cells. The most commonly used is the alpha-lectin expression system. Yeast display antibody discovery refers to the fusion of antibody sequence variable region and Aga2p expression, Aga2p protein subunit binds to Aga1p protein subunit fixed on yeast cell wall through two disulfide bonds, combined with flow sorting technology, the specific antibody targeting antigen can be screened out. Yeast surface display technology displays the target protein on the cell surface and endowing the yeast host with new capabilities. Combined with flow cell sorting, this technology can be used to quantitatively detect the equilibrium binding constant and dissociation kinetic properties of the target protein, as well as to improve the stability and specificity of the target protein, saving cumbersome purification steps. This technique can not only be used to quantitatively detect the equilibrium binding constant and dissociation kinetic properties of the target protein, but also can be used to improve the stability and specificity of the target protein, saving the tedious purification steps.
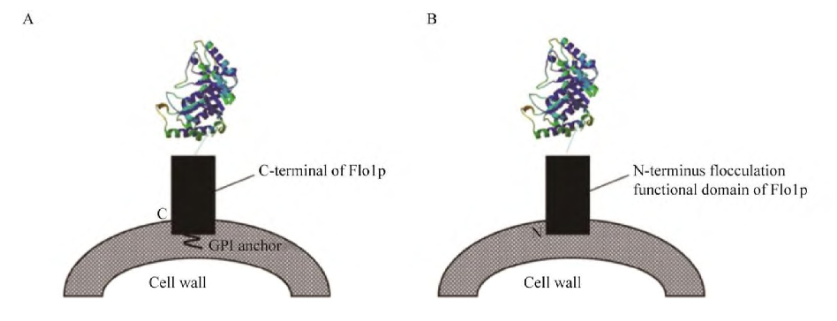
Figure 1: Schematic diagram of a surface display system for Saccharomyces cerevisiae based on α-agglutinin. A:The protein of interest (POI) is fused to C-terminal of the Aga2p with two epitope tags,HA tag and c-myc tag. B:The POI displaying on the surface of S.cerevisiae could be detected by anti-HA and anti-c-mylantibodies.
Yeast Display Library Construction Service
Yeast display libraries are a genetic engineering technique that utilizes the yeast cell surface to display exogenous proteins or peptides. Efficient presentation of exogenous proteins is achieved by linking target proteins or peptides to yeast surface proteins so that they can be presented directly on the yeast cell surface.
The key steps in constructing a yeast display library include inserting the target gene into the coding sequence of yeast surface proteins and transfecting it into yeast cells using an appropriate expression vector. Subsequently, a yeast display library is formed by screening surface display proteins with desired properties through appropriate screening methods. In antibody screening, yeast display libraries can be used to display antigen or antibody libraries, thus realizing the screening of antigen-antibody interactions. Using this technology, researchers can rapidly screen antibodies with high affinity and specificity for therapeutic and diagnostic applications.
The generation of yeast display libraries usually depends on random mutations of precursor proteins, which are generally obtained by error-prone PCR. The generated target gene mutation library and carrier enzyme digestion products are transformed into receptive yeast cells at the same time, and the intracellular mutant genes and carrier sequences are homologous recombination, so as to prepare yeast display protein mutation libraries with the size of about 10^7-10^9.
Antibodies are widely used in basic research, diagnosis, therapy and public health, and their affinity is important for the specific recognition of biomolecules. Early use of animal immunity to prepare antibodies, there are many defects, long time, high cost and difficult to obtain, the tolerance of self antigen is not high. Compared with phage display technology, yeast display technology is more suitable for the construction of antibody libraries and the screening of antibody fragments. It can distinguish proteins with twice the affinity difference, indicating that yeast display technology has higher sensitivity. VHH, with a molecular weight of about 15kDa, is an antibody with a variable heavy chain region unique to the animal from which the animal is derived. With low immunogenicity and high stability, it is easy to obtain a neutralizing antibody with high affinity, high specificity and effective resistance. It is widely used in drug target antibody development, CAR-T/CAR-NK therapy (currently widely used in CAR-T/CAR-NK therapeutic antibody). It mainly has natural advantages in the aspects of camel source VHH antibody (also known as nanobody). Compared with phage display library technology, yeast display library has a higher antibody activity level after a small amount of antibody expression. Based on this, the affinity of antibodies can be distinguished during flow screening.
Yeast Display Library Construction Service Workflow
| Steps | Detailed Introduction | Delivery Standards | TimeLine |
|
Step1: Antigen preparation |
*Antigen type: (1) Recombinant protein preparation (2) Small molecule (modified) + coupling (3) Peptide synthesis + coupling (4) Inactivated virus provided by customer (5) Customer provides encapsulated mRNA |
Antigen QC standard: Recombinant protein 3-3.5mg (purity >85%). Small molecule purity >90%. Peptide purity >90%. |
4-6 weeks |
|
Step2: Animal immunization |
(1) Animals were immunized for 4 times, with one shot of booster immunization, totaling 5 shots. (2) Collect negative serum before immunization, and collect blood in the 4th injection for ELISA to detect serum potency. (3) If the serum antibody potency of the 4th needle meets the requirements, then the blood will be collected 7 days before strengthening immunization once again, if it does not meet the requirements, then continue the routine immunization. (4) If the validity of the serum antibody meets the requirements, blood will be collected to isolate mononuclear cells. |
Animal: clear background (age, sex, immunization status). Immunization: protein/viral antigen potency >10^5, peptide/small molecule antigen potency >10^4 |
10 weeks |
|
Step3: Template cDNA preparation |
(1) PBMC total RNA extraction (RNA extraction kit) (2) High-fidelity RT-PCR preparation of cDNA (Reverse Transcription Kit) |
cDNA: uniform gel map distribution |
1 day |
|
Step4: Antibody yeast display library construction |
(1) Two rounds of PCR to amplify VHH gene using library cDNA as template. (2) Yeast display vector construction and transformation: VHH gene splicing yeast display vector, electroshock transformation of yeast, antibody library construction (3) Identification: 48 clones were randomly selected, and PCR was used to identify the positive rate; |
Sequencing was used to calculate the correct insertion rate and library diversity Library insertion rate: >90% Library Diversity: >90% Library size: 10^7-10^8 Positive library rate: >90% |
3-4 weeks |
Advantages of Yeast Display Library Construction Service
.png)
FAQ- Yeast Display Library Construction
FAQ-Yeast Display Library Construction-Gateway Technology
1. What is the yeast Library Construction (Gateway Technology) service?
A: Constructing synthetic genetic networks requires the assembly of DNA fragments encoding functional biological parts in a defined order. However, this can become a time-consuming process. To address this technical bottleneck, we created a series of Gateway shuttle and integration vectors that facilitate the assembly of artificial genes and their expression in the budding yeast Saccharomyces cerevisiae. Our method enables the rapid construction of artificial genes from promoters and open reading frame (ORF) cassettes in one-step recombination reactions in vitro. Furthermore, such produced plasmids can be easily introduced into yeast cells to test the function of the assembled genes. This system assembled DNA bait sequences and ORF libraries encoding reporter genes (His3 and Inline graphic-galactosidase) in a one-step Gateway recombination reaction. The cloned plasmids were then integrated into the HIS 3 and URA 3 loci for selection. However, the Gateway vector for the one-hybrid system was not designed as a universal plasmid vector to construct artificial genes more generally and introduce them into yeast. Such a vector should facilitate the rapid characterization of new promoter-ORF combinations before the synthesis of artificial genetic networks in yeast. The Gateway system has become a powerful high-throughput cloning method capable of in vitro recombination of DNA in a high-speed, accurate, and reliable manner.
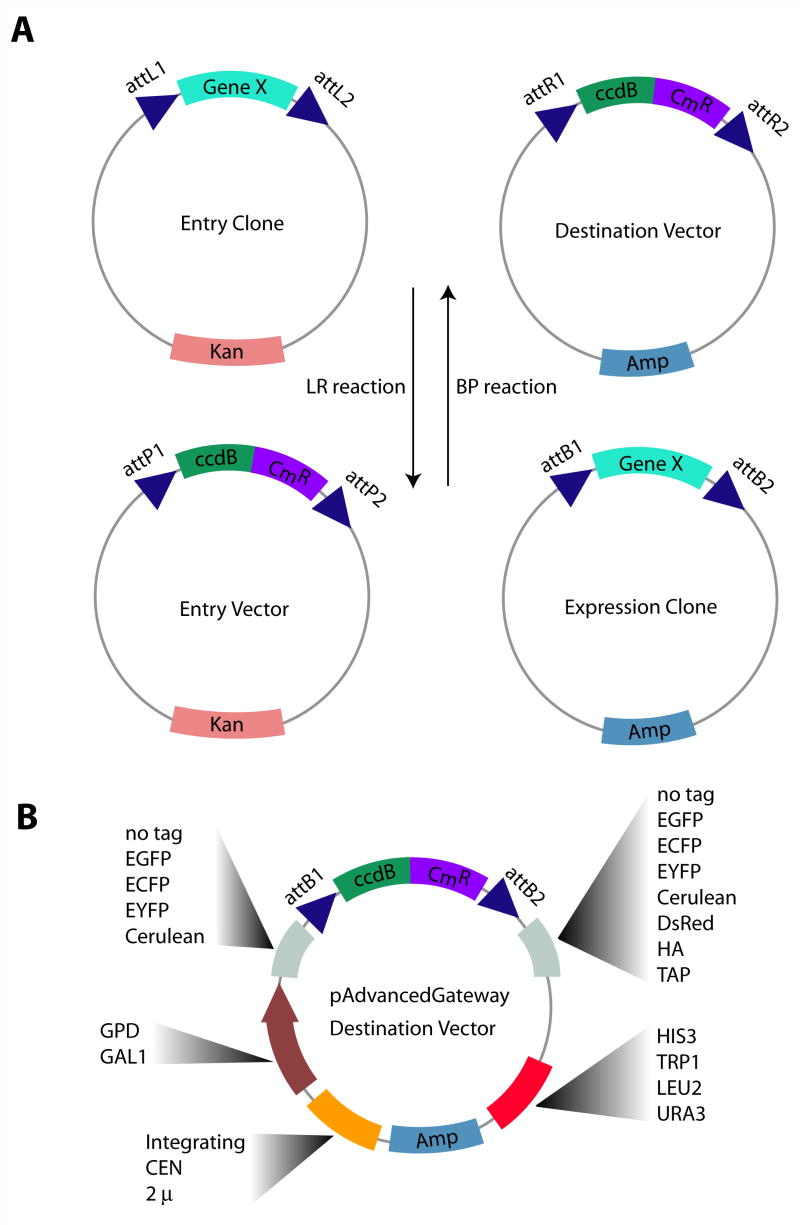
Figure 2: A Schematic representation of the construction of the yeast library. (Reference source: Alberti S, Gitler AD, Lindquist S. A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast. 2007 Oct;24(10):913-9. )
2. What are the types of Gateway technical reaction types? What is the difference?
A: There are two main types of Gateway reactions: LR and BP. The LR reaction consisted of the 'entry clone' plasmid containing the kanamycin resistance cassette (containing target genes flanking attL1 and attL2 sequences) and the target vector, containing the ampicillin resistance cassette (containing the bacterial death gene and chloramphenicol resistance gene, flanked by attR1 and attR2 sequences). The LR Clonase II Enzyme mixture (Invitrogen) catalyzes recombination between recognition sites to generate "expression clones" containing the target gene in the target vector skeleton. Transformation of E. coli with this reaction mixture and paving on LB ampicillin-containing ampicillin plates allowed a specific selection of expression clones and selective killing of bacteria containing the initial plasmid constructs. The BP response is essentially the opposite reaction to the LR, transferring the gene from the expression clone to the entry vector (isolated by screening on kanamycin-containing agar plates). The BP reaction is the first step of the Gateway technique and provides the basis for subsequent steps. It enables the convenient insertion of target genes into the entry vector, facilitating subsequent manipulation.Gateway. The LR reactions and BP reactions of the techniques varied significantly in all aspects, the two steps form the core of the Gateway technology, making it an efficient and versatile method for cloning and subcloning DNA sequences.
3. What are the advantages and applications of the Gateway technology?
A: Multisite Gateway technology enables efficient assembly of promoter and ORF in one-step reaction in vitro. The expression vectors generated by this method can be easily introduced in yeast for the functional determination of assembled genes. Similar combinatorial approaches proved to be effective for generating different phenotypes of synthetic networks in E. coli. Several different tag sequences are easily available and can be easily introduced to obtain a translational fusion because the att site does not interfere with the reading frame. The above target vector pDEST375 allows stable single-copy integration of artificial genes at the MET15 locus, adding flexible selection to the construction of artificial gene networks in yeast. Single-segment pDEST vectors can be used in almost all common systems, for example, Drosophila.Moreover, extensive pDEST libraries have been created specifically for the 2- and 3-segment MultiSite GatewayTM systems in plants. The target vector of the three-stage MultiSite GatewayTM can also accommodate gram-positive bacteria. For Saccharomyces cerevisiae, a single-segment pDEST vector is offered for yeast two-hybrid screening by Invitrogen, while a double-segment (promoter: ORF) MultiSite GatewayTM vector has been documented. Gateway Technology boasts high efficiency, versatility, flexibility, reliability, and streamlined experimental procedures, offering broad application potential in gene function research, protein expression and purification, the construction of intricate libraries, synthetic biology, as well as gene therapy and transgenic crop development.
4. What is the principle of the Gateway technology?
A: GatewayTM technology, leveraging the site-specific recombination mechanics of the phage lambda system, enables the seamless integration of phage DNA into the Escherichia coli genome.In a standard vector (pENTR), DNA fragments flanked by appropriate recombination sites can be easily transferred to a compatible vector (pDEST) for functional analysis. MultiSite GatewayTM Using the modified recombination site allows the combination of multiple DNA fragments in a separate in vitro recombination reaction. These fragments are connected in pDEST in predefined order and orientation, maintaining the reading frame, and have low mutation risk. Three-section MultiSite GatewayTM can easily make arbitrary combinations of promoters, genes, and tags without the redesign of new target vectors for each new experimental approach. The ability to select promoters allows different temporal, spatial, and quantitative control of gene expression, while different possible tags can contain fluorescent protein tags for localization or an epitope tag for detection or purification, or create protein chimeras. Multiple research groups have provided a large number of "building modules" in the form of pENTR clones, demonstrating the flexibility brought about by MultiSite GatewayTM technology. The current collection of pENTR sets encompasses ORFeomes derived from a wide array of prokaryotic and eukaryotic species. KMD Bioscience can provide customers with quality advance payment yeast library construction services.
5. In the yeast library construction (Gateway technology), what problems will be encountered, and how to solve them?
A: During the library construction process, because the Gateway technology involves two recombinations, the shaking bacteria amplification process of the primary library is similar to the PCR process, which may lead to the redundancy of high abundance genes and the loss of low-abundance genes, thus affecting the library quality. The presence of an empty vector (i.e., no vector without the target gene) will occupy the library capacity and reduce the effectiveness and screening efficiency of the library. Transformation efficiency is one of the key factors that affect the success rate of library construction. If the conversion efficiency is low, the library capacity will be insufficient, which will affect the subsequent experiments. Invitrogen Although there are many kinds of Gateway carriers provided by companies, the choice is still limited relative to traditional methods. This may have limited the need for certain specific experiments. To reduce the loss of low-abundance genes and the redundancy of high-abundance genes, KMD Bioscience optimized the recombination conditions, such as adjusting the time of the recombination reaction, temperature, and reagent concentration. In addition, more advanced screening techniques, such as high-throughput sequencing, can also be used to assess the library's quality and diversity. During the construction of the library, quality should be strictly controlled, including the use of high-quality reagents, optimizing operation steps strengthening monitoring of quality control points, etc.
FAQ-Yeast Display Library Construction-SMART Technology
1. What is the yeast library construction? What does the SMART technology mean?
A: In the past two decades, yeast display technology has become a key technology to explore and optimize protein stability, and has been widely used in various fields, among which in the heterodimeric proteins such as antibody and T cell receptor (TCR) screening, yeast surface display library technology shows its unique advantages, because the yeast surface display library can use the mechanism of gap repair to complete the drive of homologous recombination technology, and then help us to build the library, and can further build a combined library after the fusion of different kinds of yeast, which greatly enriches the diversity and efficiency of screening. The yeast library construction as the name implies is a method for constructing gene libraries using yeast as an expression vector. Yeast libraries are composed of many gene clones that, when inserted into vectors, can replicate and express within yeast cells. SMART amplification extracted total RNA from organisms and performed cDNA synthesis using the conversion mechanism (SMART) at the 5' end of the RNA template. Subsequently, the double-stranded cDNA was amplified by long-distance PCR, purified in the yeast strains, and co-transformed with the vector. The quality parameters of the constructed libraries were evaluated to verify the constructed libraries. KMD Bioscience can build yeast libraries for customers.
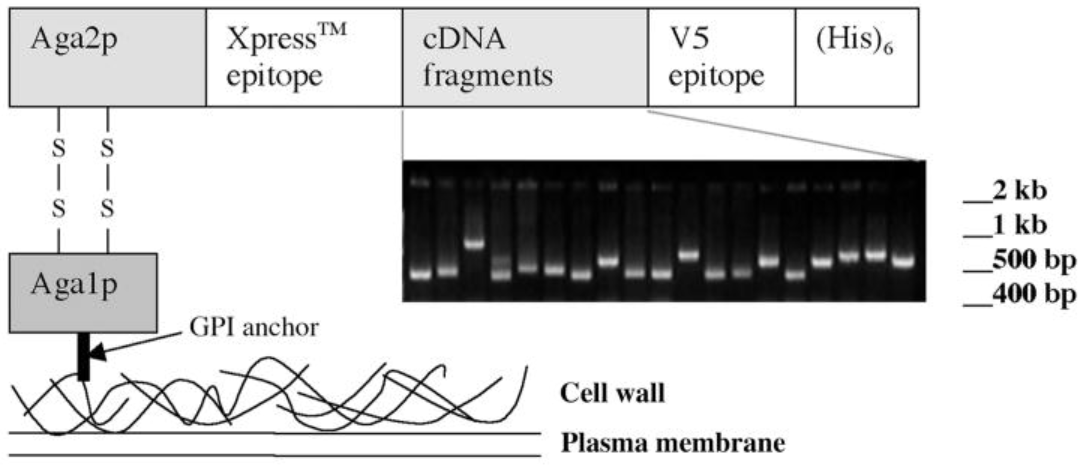
Figure 3: Yeast Display Library Construction. (Reference source: Bidlingmaier S, Liu B. Construction of yeast surface-displayed cDNA libraries. Methods Mol Biol. 2011;729:199-210. )
2. What are the advantages of SMART technology? What is the general application?
A: SMART technology can complete the amplification of full-length cDNA with high quality and efficiency even if the initial sample RNA is small. This technology is more suitable for the cloning of low-abundance expressed genes. After homogenization with some technical means, we found that SMART technology significantly reduced the redundancy of genes, which helped us to effectively enrich those genes with lower expression levels, which could improve the gene diversity in our constructed yeast library. Compared with other technologies for building yeast libraries, SMART technology has a simpler operation process and faster operation, which is conducive to researchers to quickly carry out experiments in the laboratory. SMART technology utilizes the template switching activity of the retromer, adding a specific sequence to the 5' end of the cDNA, which could increase the efficiency of the full-length clone. KMD Bioscience has a strict quality control process. Through the quality control process, the yeast library constructed by SMART technology can reach high standards in terms of library capacity, average insert length, and clone positive rate. Researchers at KMD Bioscience can build high quality and high reliability yeast libraries, providing strong support for in-depth research in different life sciences.
3. How does SMART require RNA initiation in the yeast library construction service?
A: One of the significant advantages of SMART technology in the field of molecular biology is that it is highly inclusive of RNA initiation. In experiments, the technology requires only a very low starting amount of RNA, usually only a few micrograms (µg) of high-quality RNA, to start and complete the complex library-building process. This feature is crucial for projects where RNA samples are difficult to obtain, such as our common rare cell type analysis, low-abundance gene expression studies, or limited clinical sample resources in hospitals. In this case, the SMART technology can greatly reduce the threshold of the experiment, without making our experiment limited by the sample size. Despite the low sample size required by SMART technology, we should pay attention to the quality of the RNA samples. The quality of the RNA samples has a significant impact on both the quality and diversity of the final constructed libraries. High-quality RNA ensures that the experiment reduces biases and errors during transactivation and amplification, thus reflecting the true situation of the original gene expression. Therefore, in the case of SMART technology, strict RNA extraction and purification methods are also necessary to ensure that pure, intact, and not degraded RNA samples are obtained and that the RNA samples do not affect subsequent experiments.
4. How efficient is SMART technology to build yeast libraries? How to ensure the diversity of yeast libraries constructed by SMART technology? How can we solve the above problem?
A: SMART introduces a switching reaction at the 5'end of RNA through its unique amplification mechanism. This technology can not only retain the original information of the RNA template but also help achieve efficient amplification of cDNA fragments. However, SMART may not be as efficient in some aspects, as some other library building techniques Gateway recombination than others. In the face of these technical shortcomings, tech biological researchers actively explore and the optimization of the experimental conditions, and experimental scheme optimization including but not limited to adjusting the proportion of the RNA extraction solvent and temperature control in the extraction, the selection and amount of retrospection used in the optimization process, control the reaction time, cycle number and the pH value in the optimization reaction system. All of these schemes mentioned above can help us to improve the efficiency of library construction of SMART technology. When we were faced with the problem of insufficient library diversity, we found that the key to ensuring library diversity lies in the diversity and integrity of the starting RNA samples. When extracting RNA, we should try to avoid the degradation and contamination of RNA to ensure the integrity of RNA. During library construction, we should also pay attention to controlling the bias of reverse transcript and amplification and help reduce the over-amplification of specific sequences, which can help us improve the diversity of yeast libraries.
5. In the yeast library construction service, how do we evaluate the quality of yeast libraries constructed by SMART technology? False positive yeast library constructed by SMART technology? How do we solve it?
A: In the yeast library construction service, how well the quality of the yeast library constructed by SMART technology is directly related to the effectiveness and accuracy of subsequent studies. We can evaluate the library quality through the following several aspects. The first is library capacity, which can directly reflect the degree of genetic diversity contained in libraries, and high-capacity libraries can provide a wider range of genetic resources for screening and study. Then there is the size of the insert, which is related to the gene segment in the library, and the most successful insert that includes the full length or the critical region of the target gene. Also, how many positive clones are in a library can be assessed for quality. Positive clones can show how many clones correctly insert the target gene fragment, and high positive clones indicate a high screening and success rate. In addition to the above indicators, the validation of the functionality can also evaluate the library quality. Meanwhile, the false positive problem also often appears in library construction and is not limited to SMART technology. In the face of false positives, to reduce false positives, we can add more stringent quality control measures in the process of library construction and screening, strictly control various experimental conditions, and conduct multiple verification and bioinformatics analyses of the results.
How to Order?
If you have any questions regarding our services or products, please feel free to contact us by E-mail: [email protected] or Tel: +86-400-621-6806