2024-10-11 Hits(153)
Peptide Library Screening
In 1985, Smith genetically engineered filamentous bacteriophages and obtained recombinant bacteriophages that could proliferate in vitro, creating phage display technology. In 1988, the idea of constructing a phage peptide library to understand antigenic epitopes was proposed by fusing the N-terminus of the pIII protein of bacteriophages with known antigenic determinants and presenting them on the surface of bacteriophages, which enabled the fusion to specifically bind to antibodies. As a technology for studying antigenic epitopes, phage display mainly involves inserting exogenous genes into phage capsid proteins through genetic engineering techniques, so that the peptides encoded by exogenous genes can be displayed as fusion proteins without changing their own structure and activity. Then, through multiple rounds of screening, highly specific antibodies or peptides that can bind to the target molecule are selected. In 1990, a random 6-peptide library based on phage display technology was created. After years of development, phage display technology has been applied not only in the study of antigen epitopes, but also in areas such as cell signal transduction, protein recognition sites, and drug development. The peptide library established based on phage display technology has a capacity of billions. In terms of screening peptide markers, phage display peptide libraries have the advantages of speed, relatively low cost, and large library capacity. By screening bacteriophage peptides that bind to specific antigens, new antibodies can be developed or existing antibodies can be improved. Bacteriophage displayed peptides can also be used for vaccine development, cancer diagnosis, biosensor development, and more.
Classification of peptide libraries
Peptide libraries based on phage display are mainly composed of short peptide sequences of different lengths, which are classified into different types according to different construction methods and applications. Overlapping peptide library is a phage peptide library constructed by gradually cutting different fragments of protein sequences. It is mainly used for full-length proteins and helps to recognize continuous linear epitopes. The peptide library constructed by removing the amino acids on both sides of the original peptide sequence is called a truncated peptide library, which is used for the determination of small epitopes and helps to identify amino acid sequences related to biological activity. In the alanine peptide scanning library, alanine can replace every amino acid residue to understand the role played by each amino acid. The random peptide library contains combinations of peptides with all possible arrangements of 20 amino acids. Due to the randomness of these peptide sequences, the random peptide library contains a large number of different peptides, which can be used to discover more peptides with specific functions or interactions. Disordered peptide libraries can rearrange and combine existing amino acid sequences with high variability, and are often used as negative controls to demonstrate the importance of amino acid sequences arranged in a specific order for the structure and function of peptides.
A peptide library composed of amino acids arranged linearly is called a linear peptide library. It has a simple structure, is easy to synthesize and characterize, and can adjust the sequence and length of amino acids as needed. It is mainly used to study protein-protein interactions and screen antigenic epitopes. Some chemical reactions, such as amide ring bonding, can transform linear peptide libraries into peptide chain libraries with cyclic structures, known as cyclic peptide libraries. Due to the ability of cyclization to promote the binding of intramolecular hydrogen bonds, reduce the ability of external hydrogen bonds, improve protein resistance, and prolong half-life, cyclic peptide libraries have stronger stability. Due to the relatively large surface area of cyclic peptides, the probability of binding to receptors increases, and they can even target proteins without binding pockets. Different cyclization methods and binding sites can generate a large number of cyclic peptide molecules, increasing the possibility of discovering new ligands. Some cyclic peptides have strong cell penetration and oral properties, and have potential application value in the development of therapeutic drugs.
Synthesis and screening of peptide libraries
The basic principle of constructing a phage peptide library is to create a library containing a large number of peptide sequences of different lengths, amino acid compositions, and structures, and then identify peptides with specific functions through random peptide library screening. The synthesis steps include the generation of random peptide sequences, the synthesis of peptides, and their display or expression on specific carriers. When constructing a 12 peptide library, the algorithm is first used to generate a large number of random 12 peptide sequences. To ensure the diversity of the library, these sequences can cover all possible amino acid combinations. Afterwards, the synthesis of peptides is carried out using solid-phase synthesis, which is currently the most commonly used method. In order to prevent side reactions, all amino acids involved in the reaction are protected by protective groups, such as Fmoc and BOC. First, the C-terminal amino acid is connected to the solid-phase carrier, and its protective group is removed. The remaining amino acids are added sequentially, and the steps of washing out the protective group, connecting, and washing are repeated until the complete 12 peptide sequence is synthesized. The generated 12 peptide sequence was fused to the N-terminus of the M13 phage pIII protein, and then screened and identified using phage display technology.
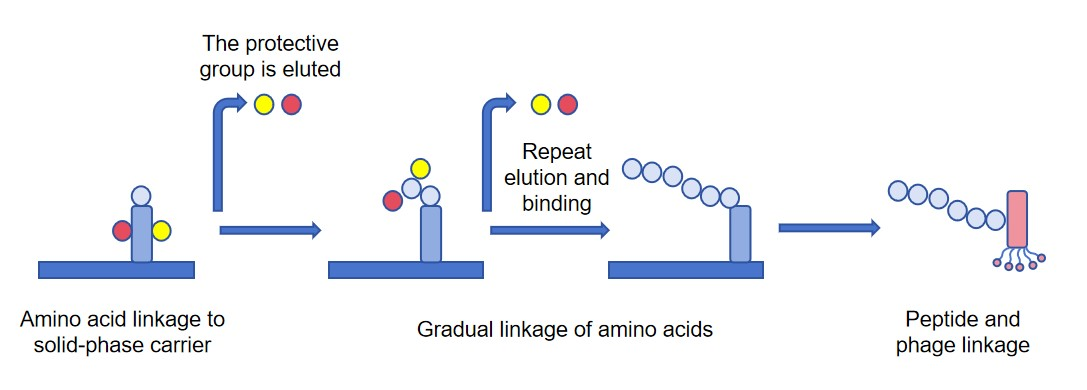
Fig. 1 Schematic diagram of peptide library synthesis
Solid phase screening and liquid phase screening are two commonly used methods for random peptide library screening. Solid phase screening involves immobilizing target molecules on a solid phase carrier, such as a 96 well plate. The peptide library is then incubated with the immobilized target molecules, and unbound peptides are removed by washing. Finally, bound peptides are eluted from the solid phase carrier for subsequent analysis. The operation of solid-phase screening is relatively simple, without the need for antigen labeling. It can process multiple samples simultaneously and is suitable for large-scale screening, but it is also prone to non-specific binding. Liquid phase screening mainly uses magnetic beads and other screening platforms to bind target antigens to magnetic beads, incubate them with peptide libraries, and then separate the magnetic beads bound to peptides for subsequent analysis and identification. Liquid phase screening can reduce the occurrence of non-specific binding, but requires special equipment to operate, suitable for screening with reduced antigen concentration gradients and screening of high affinity peptides.
KMD Bioscience is committed to providing customers with high-quality peptide library construction technology services. We have rich project experience and insights in peptide library construction. After years of development, KMD Bioscience has established a complete peptide library construction system, including M13 phage display system, M13 KE phage display system, T7 phage display system, etc. that rely on M13 auxiliary bacteriophages. We can provide customers with high-quality linear peptide libraries (including but not limited to 6-peptide library, 7-peptide library, 12 peptide library, 15 peptide library) and various types of peptide library construction services such as cyclic peptide libraries (cyclic 6-peptide library, cyclic 7-peptide library, cyclic 10 peptide library, etc.). The M13 phage peptide library established by KMD Bioscience has a storage capacity of up to 108 and a titer of up to 1013 phage displayed peptide particles/ml. The T7 phage peptide library has a storage capacity of up to 108 and a titer of up to 1011 phage displayed peptide particles/ml, which is sufficient to support customers in screening targeted peptides for various targets and meeting downstream experimental needs in the future. KMD Bioscience can provide one-stop technical services including peptide gene library design and synthesis, peptide library construction, matching peptide library screening, affinity validation, in vitro cell validation, etc. Customers only need to provide specific project requirements, and KMD Bioscience scientists will design the best library construction method and phage system based on customer project requirements to meet their project needs.
Reference
[1] Jaroszewicz W, Morcinek-Orłowska J, Pierzynowska K, et al. Phage display and other peptide display technologies. FEMS Microbiol Rev. 2022;46(2):fuab052.
[2] Chen PC, Liu WR. The Construction of a Genetically Encoded, Phage-Displayed Cyclic-Peptide Library. Methods Mol Biol. 2021;2355:219-230.
[3] Gao Q, Chen L, Jia C, et al. Selection and identification of a specific peptide binding to ovarian cancer cells from a phage-displayed peptide library. Biotechnol Lett. 2022;44(8):951-960.