2024-07-03 Hits(346)
Antibody Discovery
Introduction of Phage Display Technology
Phage display technology is an in vitro antibody screening method developed by George P. Smith in 1985. Phage display technology is an in vitro antibody screening method developed by George P. Smith in 1985. Phage display technology displays peptides, proteins, antibodies, etc. on the surface of bacteriophages. This method has become an effective method for producing large quantities of peptides, proteins, and antibodies, allowing for the construction of libraries containing up to 10^10 different variants. It can be used for affinity screening of peptide libraries to study protein-ligand interactions, antigens, and antibody binding sites, and the production of monoclonal antibodies.
The Principles of Phage Display Technology
The principle of phage display technology is to insert genes encoding exogenous peptides or proteins into the appropriate positions of the bacteriophage's outer shell protein structure genes. With normal reading frames and without affecting the normal function of the outer shell protein, the exogenous peptides or proteins form fusion proteins on the bacteriophage's outer shell protein, which is presented on the surface of the bacteriophage as the offspring bacteriophage reassembles. The displayed protein can maintain a relatively independent spatial structure and biological activity, which is conducive to the binding of target proteins. Therefore, target proteins can be quickly used for multi-round screening of phage display antibody libraries and expanded cultivation in E. coli, a process also known as "panning". Repeated screening can gradually increase the proportion of phages that specifically recognize target molecules, ultimately obtaining peptides or proteins that recognize target molecules.
The Process of Phage Display Technology
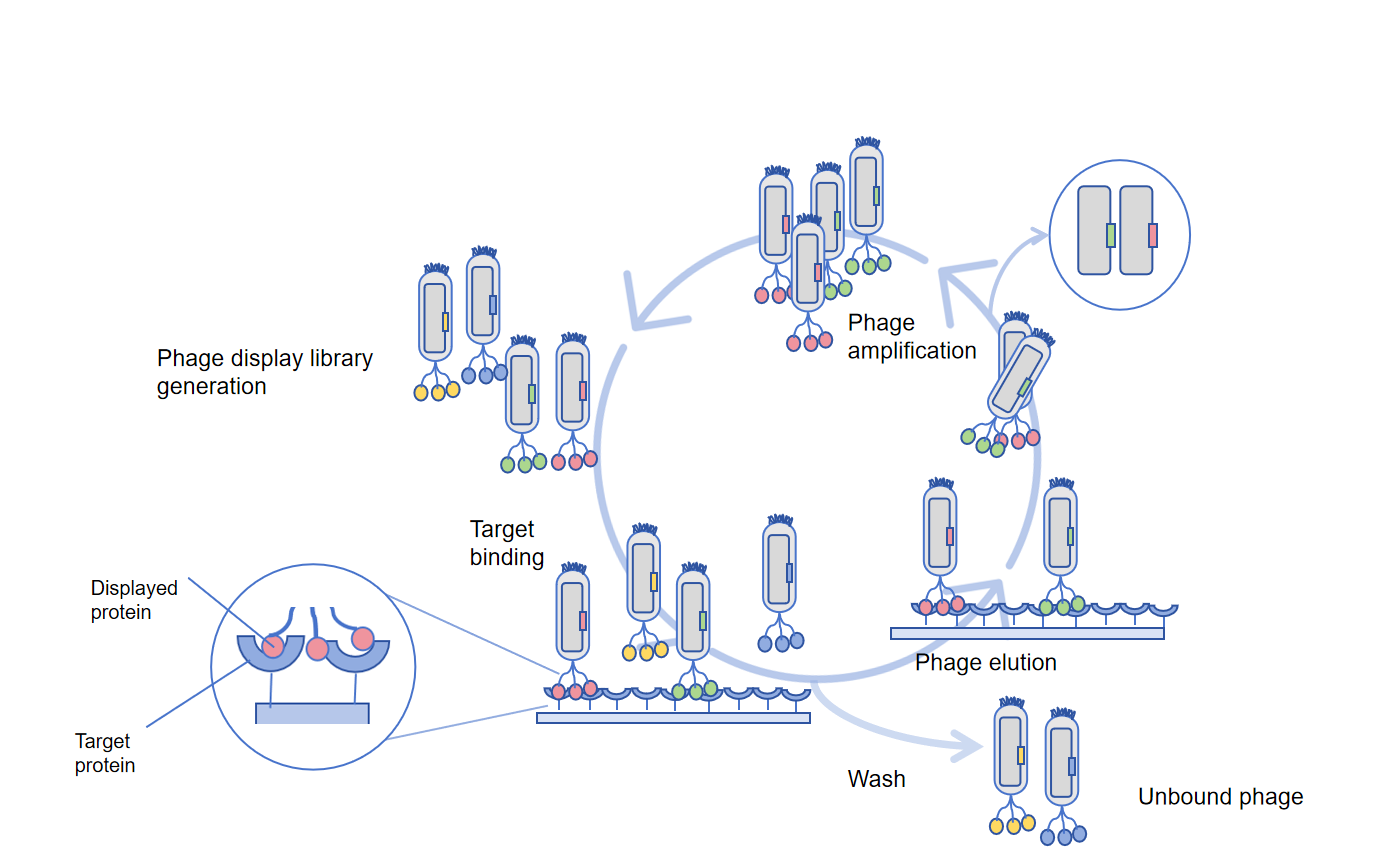
Phage display technology is a high-throughput method used for antibody discovery to different antigens. Phage display screening steps for phage display technology:
Fix the target.
Target molecules (peptides, proteins, etc.) can be immobilized
Adsorb onto a 96-well plate, rinse off unbound target molecules and seal with unrelated proteins or non-ionic detergents.
Phage binding.
Add a phage library with a storage capacity of 10^6-10^11, and add the library to the 96 well for antigen presentation for binding. The phage-binding antigen that displays the highest affinity antibody is then washed to remove non-binding phages.
Elution.
After washing, the bound bacteriophages are washed off by washing with dilute acid or alkali, due to the stability of bacteriophages, extreme pH, denaturants, ion strength, limited protein hydrolysis, or ultrasound treatment can all be used for non-specific elution of phage.
Amplification.
Add the M13 helper phage and amplify the washed high-affinity phage in Escherichia coli. To obtain high-yield and high-affinity antibody fragments, these four steps are usually repeated 3-6 times for amplified phages from the previous round, increasing the selection round to isolate phages with higher affinity. The final titer and ELISA determination of bacteriophages are used to confirm the selection of binding, followed by selecting positive bacteriophage plaques for cultivation and analyzing the sequence through NGS sequencing.
Phage display technology uses antibody libraries, which can be permanently preserved once built without the need for repeated immunization and phage display library construction. This allows for repeated screening, and after screening, there is no need to clone each antibody gene separately, allowing for direct molecular transformation. In addition, phage display technology can target a wide range of antigens. Compared with other display technologies such as ribosomes, yeast, or mammalian cell displays, one of the advantages of phage display is the ability to create large libraries (>1011 monoclonal diversity), promoting the discovery of high-affinity antibodies targeting a wide range of antigens. However, due to the inability of bacteriophages to infect excessively long sequences, additional "combination" steps need to be added. Occasionally, the combined monoclonal antibodies may lose affinity or produce autoimmunity, and phage display technology antibodies have not undergone in vivo maturation, resulting in random pairing of heavy and light chains and limited affinity.
So far, phage display technology has developed relatively maturely and has been widely used in antigen epitope analysis, protein-protein interaction, enzyme specificity and inhibitors, antibody development, transfusion medicine, Diagnostic and therapeutic agents for autoimmune diseases, monoclonal antibodies production, antibody humanization, drug screening, vaccine development, immunological disease diagnosis, and so on.
Reference
[1] Ledsgaard L, Kilstrup M, Karatt-Vellatt A, McCafferty J, Laustsen AH. Basics of Antibody Phage Display Technology. Toxins (Basel). 2018 Jun 9;10(6):236. doi: 10.3390/toxins10060236. PMID: 29890762; PMCID: PMC6024766.
[2] Bazan J, Całkosiński I, Gamian A. Phage display--a powerful technique for immunotherapy: 1. Introduction and potential of therapeutic applications. Hum Vaccin Immunother. 2012 Dec 1;8(12):1817-28. doi: 10.4161/hv.21703. Epub 2012 Aug 21. PMID: 22906939; PMCID: PMC3656071.
[3] Jyoti Pande, Magdalena M. Szewczyk, Ashok K. Grover. Phage display: Concept, innovations, applications and future, Biotechnology Advances, Volume 28, Issue 6,2010, Pages 849-858, ISSN 0734-9750