2025-02-14 Hits(259)
CO-IP
Co-IP assays are similar to immunoprecipitation assays in that they are both proteins that detect protein-protein interactions. Co-IP technology is used to analyze whether there is an interaction between two or more proteins, based on immunoprecipitation experiments, Compared with other protein-protein interaction experimental techniques (GST Pull-down, Western Blot, etc.), Co-IP technology has higher specificity, sensitivity, and high repeatability, which can avoid the interference of non-specific binding and reflect the interaction of proteins in the natural state.
Co-IP assays have the following advantages: 1. It can provide evidence directly for protein-protein interactions that enable precise targeting of interacting proteins, and direct capture of specific protein complexes from cell lysates. 2. To ensure the natural state of the protein complex, and to provide more real biological information for research. 3. It can provide great flexibility and a wide range of research applications that Can be applied to specific tissue cells or whole organisms.
Co-IP assays are currently used in disease mechanism research, drug development, biomarker discovery, etc. It provides a new target for disease diagnosis and treatment, and the interaction of disease-related proteins can be revealed; Understanding the molecular mechanism of drug action accelerates the process of new drug development; It provides the possibility for early diagnosis of diseases that plays a key role in the discovery of biomarkers.
Principle
The principle of the Co-IP assays is to extract the protein from the experimental sample and then add things in turn of specific antibodies and protein A/G magnetic beads. These above things incubate for some time which magnetic beads and the antiboy-protein complex need to bind for enough time. The supernatant of the unbound protein is discarded by adsorption of the magnetic bead on the magnetic rack, and the magnetic bead is washed cycle that removes the protein and other impurities unless specifically bound. The antibody-protein complex was obtained by elution of decomplex buffer, which can be used for subsequent silver staining, immunoblotting, and mass spectrometry experiments.

Fig.1 Schematic diagram of Co-IP assays
Samples
Co-IP experimental protein samples are usually obtained from plenty of cracking samples, including various plant and animal tissue samples, bacteria, cells, etc. When the protein abundance of the sample is low or the interaction force is not strong, the experiment failure is easy to occur. The solution is to increase the protein concentration of the sample which can maintain the original protein interaction state. Since the experimental conditions need to be explored repeatedly, there are certain requirements on the number of samples (cell > 2x107, animal tissue > 500mg, plant tissue > 2g), and the samples are stored at -80℃ after collection to avoid repeated freezing and thawing.
Pay attention to the time of cracking and whether the cracking fluid is mild and the most suitable during the processing of the sample, Whether the lysis is complete and the protein-protein complex is destroyed are the key factors affecting the success of the experiment, and the selection of specific antibodies is another key factor affecting the success of the Co-IP assays.
Technical Case Result Diagram
(1) Silver Staining Result Chart
Co-IP detects protein quality and cuts glue that sends it to the mass spectrometry service to reduce interference from miscellaneous proteins by silver staining. It has higher sensitivity and is more suitable for detecting samples with lower protein or lesser target protein content, compared with the WB assay.
The evaluation criteria of the silver staining experiment: When the IP group had more protein bands than the IgG group there were differences, indicating that Co-IP captured interacting proteins.
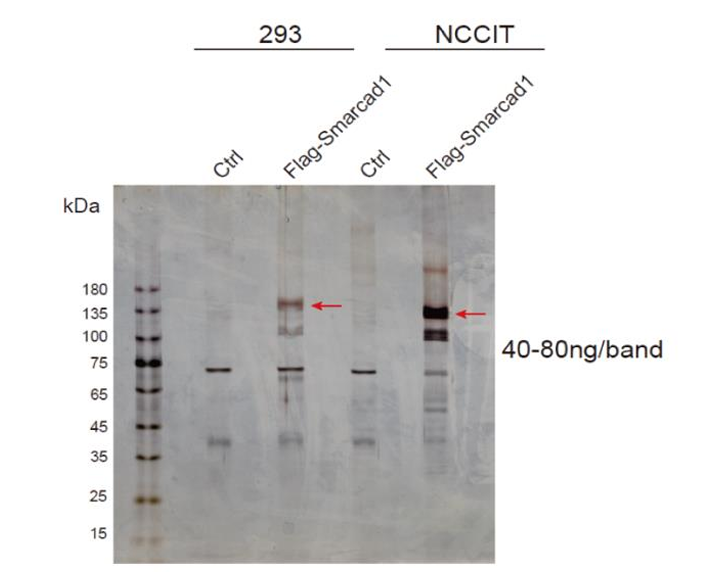
Fig 2: Silver staining result
(2) Western Bolt Result Plot
Co-IP detection can verify whether two known proteins interact with each other through WB, and can generally be used for a certain signaling pathway.
The evaluation criteria of the WB experiment were: If the IgG group had no target protein bands and the IP group had target protein bands, indicating that Co-IP captured interacting proteins.
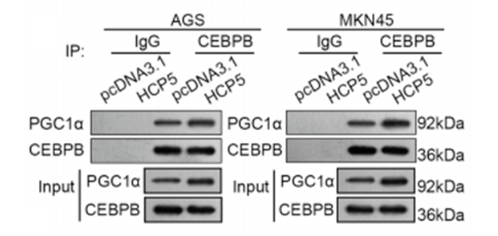
Fig 3: WB result
(3) MS Result Plot
Mass spectrometry is to enzymolize the product protein, digest and determine it into a polypeptide, and obtain the sequence. Compare it with the protein database to the results of potential binding proteins.
Co-IP assays can be used to screen and identify proteins that may bind to the protein of interest by mass spectrometry, and can also be narrowed down by two to three parallel experiments to save time in research.
The evaluation criteria of the MS experiment: The number of IP products identified by mass spectrometry has nothing to do with the result, it depends on the number of interacting proteins and the strength of the binding force of the target protein itself.
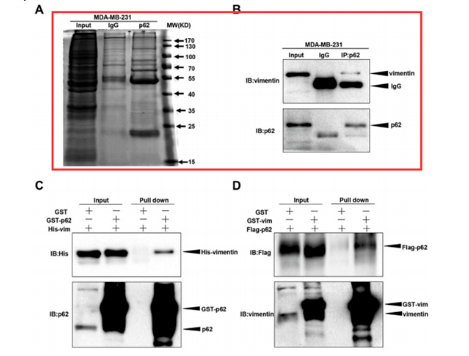
Fig 3: MS result
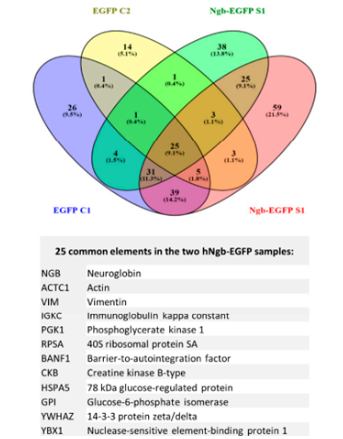
Fig 4: Co-Ip example
Based on the above, KMD Bioscience can provide various sample processing services and corresponding Co-IP assay services, compared with other biological companies, KMD Bioscience's Co-IP technology comes from the IPS laboratory of Kyoto University in Japan, which can provide high-quality experimental data and support to promote the smooth progress of your research or project.
In addition, KMD Bioscience also provides protein-protein interaction services, including yeast two-hybrid, pull-down, immunoprecipitation assays, etc., to meet the experimental needs of different customers.
References
[1] Lin JS, Lai EM. Protein-Protein Interactions: CoImmunopre- cipitation [J]. Methods Mol Biol, 2017, 1615: 211-219.
[2] Tan L, Yammani RR. Co-Immunoprecipitation-Blotting: Analysis of Protein-Protein Interactions [J]. Methods Mol Biol. 2022, 2413: 145-154.
[3] Tang Z, Takahashi Y. Analysis of Protein-Protein Interaction by Co-IP in Human Cells [J]. Methods Mol Biol. 2018, 1794: 289-296.