2025-03-14 Hits(161)
Protein Interaction
Pull-down assays (protein pull-down) are in vitro methods for determining interactions between multiple proteins and can be used to detect interactions between known and unknown proteins. The protein pull-down assays are similar to the Co-IP assays, both of which belong to the detection of protein interaction experiments. The difference between the two methods is that the Co-IP assays obtain the interacting proteins of known proteins, which has a certain authenticity but can‘t confirm whether the interacting proteins interact directly or indirectly. Pull-down assays are to find the unknown protein interacting with the known protein. It can directly confirm whether the known protein interacts with the detected protein but can’t know the binding situation in vivo.
Principle
Pull-down assays are to fix known proteins on magnetic beads to adsorb proteins that can interact, and then remove the protein complex system by changing the eluent or elution conditions. Liquids containing interacting proteins are collected, and detected by SDS-PAGE or WB assay.
In 1988, Smith et al. purified GST fusion protein, which was applied in pull-down assays and is called gst pull down. gst pull down is to add a GST tag to the target protein, capture the interacting protein through GSH, eluate the binding, and verify it through WB assays.
His Pull-down is to use metal ions such as nickel or cobalt as the medium, purify the protein through affinity chromatography, and eluate the binding through WB experiment for verification.
In addition, there are DNA Pull down, RNA Pull down, and small molecule pull down assays.
DNA Pull down is an in vitro method of determining the DNA binding of protein. Whether specific proteins bind to target DNA was detected by WB assays; Unknown DNA fragments bound to proteins, which DNA fragments are known by MS detection
RNA Pull down is an in vitro method for determining RNA binding to proteins. Whether specific proteins bind to target RNA was detected by WB assays; Unknown RNA fragments bound to proteins, which RNA fragments are known by MS detection
SM pull Down (small molecule pull down assays) can find target proteins that bind specifically to small molecules, and when combined with MS, small molecule target proteins can be accurately screened, which can be divided into the in vitro labeling method and biological orthogonal method.
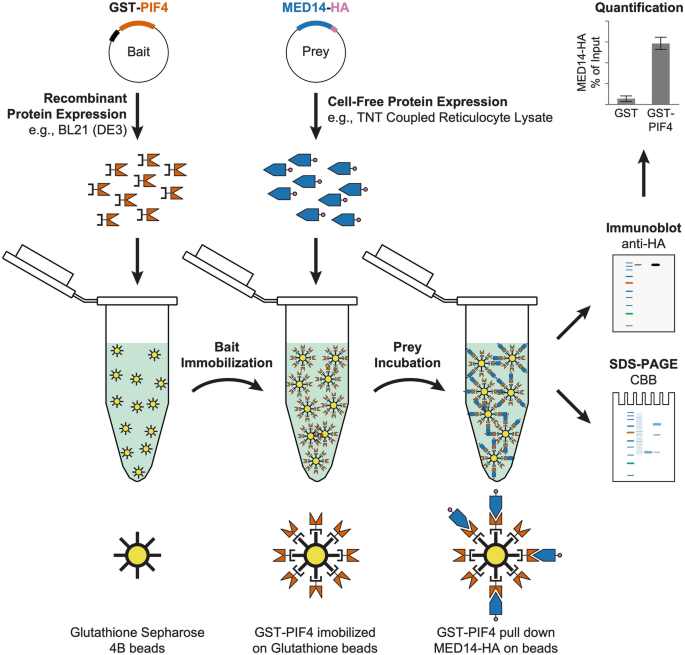
Fig. 1 Schematic diagram of gst pull down experiment
Sample
The samples come from animal and plant tissues, cells, etc., and the amount of samples can affect the content of proteins.
(1) Prokaryotic protein samples:
His fusion protein was obtained by purification of plasmids containing His label expression, and GST fusion expression protein was obtained by purification of plasmids containing GST label expression.
(2) Eukaryotic protein sample
Iysate cells to obtain cell lysate containing target proteins.
(3) Animal tissue samples
Substances such as fat were removed during sample preparation to avoid affecting the experimental results.
(4) Plant tissue samples
The abundance of chlorophyll and other substances in plant leaf tissues is high, which has a certain impact on the experimental results.
Technical Case Result Chart
(1) Silver Dyeing Results
Pull down detection can detect protein quality through silver staining, and the silver staining results are cut and sent to the mass spectrometry service to reduce the interference of miscellaneous proteins. Compared with WB, the silver staining method is more suitable for detecting samples with lower or less target protein content.
The evaluation criteria of the silver staining experiment: based on the case, it showed that the target probe group had differential protein pull-out, so subsequent mass spectrometry could be performed.
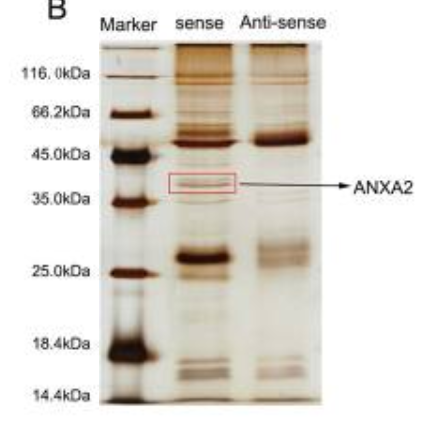
Fig 2: Silver dyeing results (Reference source:LINC00941 promotes pancreatic cancer malignancy by interacting with ANXA2 and suppressing NEDD4L-mediated degradation of ANXA2 )
(2) WB Result Chart
WB is one of the ways to verify whether two known proteins interact.
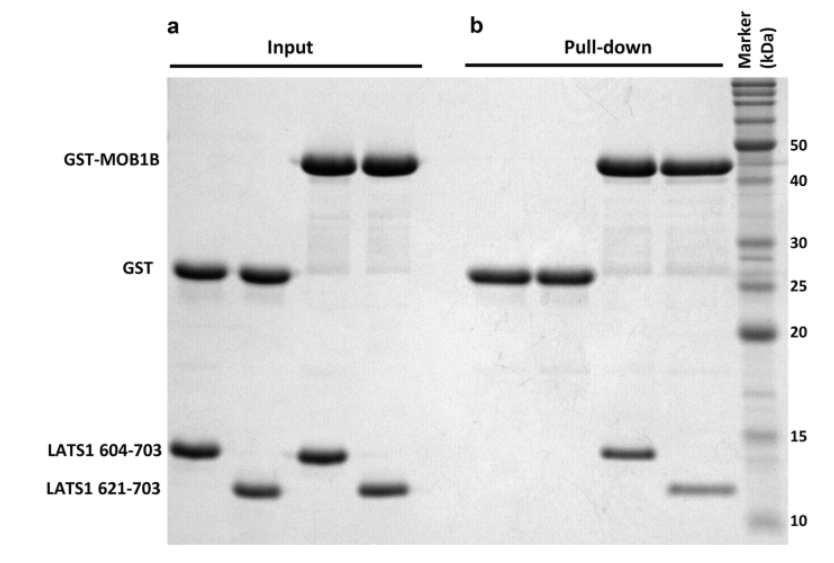
Fig 3: WB result (Reference source: GST Pull-Down Assay to Measure Complex Formations)
(3) MS Result Diagram
Mass spectrometry is to enzymolize the product protein, digest and determine it into a polypeptide, and obtain the sequence. Compare it with the protein database to the results of potential binding proteins.
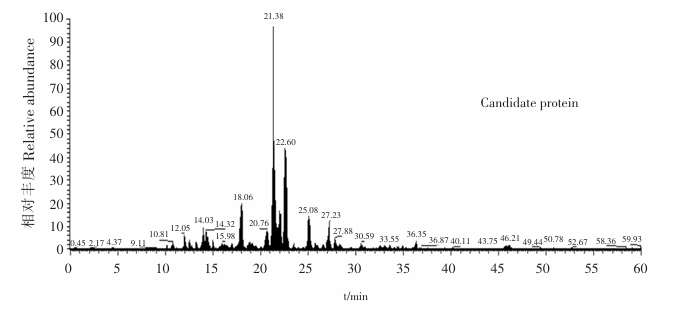
Fig 4: MS result
Based on the above content, KMD Bioscience can provide various sample processing services and corresponding Pull-down experimental services. Compared with other biological companies, KMD Bioscience Company's Pull down technology is derived from the IPS Laboratory of Kyoto University in Japan, which can provide high-quality experimental data and support to promote the smooth progress of your research or project.
In addition, KMD Bioscience also provides protein interaction experiment services, including yeast two-hybrid, Co-IP, and immunoprecipitation assays to meet the experimental needs of different customers.
References:
[1] Kim SY, Hakoshima T. GST Pull-Down Assay to Measure Complex Formations [J]. Methods Mol Biol. 2019, 1893: 273-280.
[2] Lyu S, Zhang C, Hou X, Wang A. Tag-Based Pull-Down Assay [J]. Methods Mol Biol. 2022, 2400: 105-114.
[3] Louche A, Salcedo SP, Bigot S. Protein-Protein Interactions: Pull-Down Assays [J]. Methods Mol Biol. 2017, 1615: 247-255.