2025-04-09 Hits(87)
Protein Expression
Overview of Protein
Proteins are biomacromolecules consisting of one or more chains of amino acids, the basic building blocks of proteins, which are joined by peptide bonds to form long chains. Proteins have a primary structure (linear sequence of amino acids), a secondary structure (folding patterns of local regions, such as α-helix and β-folding), a tertiary structure (three-dimensional folding of the entire polypeptide chain), and a quaternary structure (three-dimensional structure of a complex protein consisting of multiple polypeptide chains (subunits)). Proteins are the basic building blocks of life, and they play a variety of roles in the structure and function of cells, including enzymes, structural proteins, transport proteins, regulatory proteins, and immune proteins.
Natural proteins are proteins produced in nature through the normal biosynthesis process of organisms. There are thousands of different proteins in nature. Protein synthesis, folding and modification is a complex process in organisms. Organisms control protein synthesis by regulating gene expression, and natural proteins adapt to specific environments in the long-term evolution process.
Recombinant proteins are proteins synthesized through genetic engineering techniques in a heterogenous expression system, usually a gene from another organism. Recombinant proteins can be mass-produced in E. coli, yeast, or mammalian cells and purified by a variety of chromatographic techniques. Recombinant proteins are widely used in medicine, research and industry, especially in the production of therapeutic proteins and vaccines[1]. The protein production process allows for better control of pathogens and impurities, and therefore may be safer in some cases. However, recombinant proteins may not be able to fully reproduce all the properties and functions of natural proteins in their natural environment, especially in terms of post-modification such as glycosylation.
Natural proteins and recombinant proteins each have advantages and limitations, and they play an important role in different fields of application. With the development of science and technology, the production and application of recombinant protein will be more extensive and in-depth.
Recombinant Protein Expression
Recombinant protein expression is the use of DNA recombination technology to introduce target genes (foreign genes) into host cells and make them express specific proteins through the biological mechanism of host cells [2].
The basic steps to obtain recombinant proteins include: Target genes are amplified, inserted into clonal vectors, subcloned into expression vectors, transformed into protein-expressing hosts (E. coli, yeast cells, mammalian cells or baculovirus-insect cells), recombinant protein identification tests (SDS-PAGE, Western blot), protein amplification and purification [3].
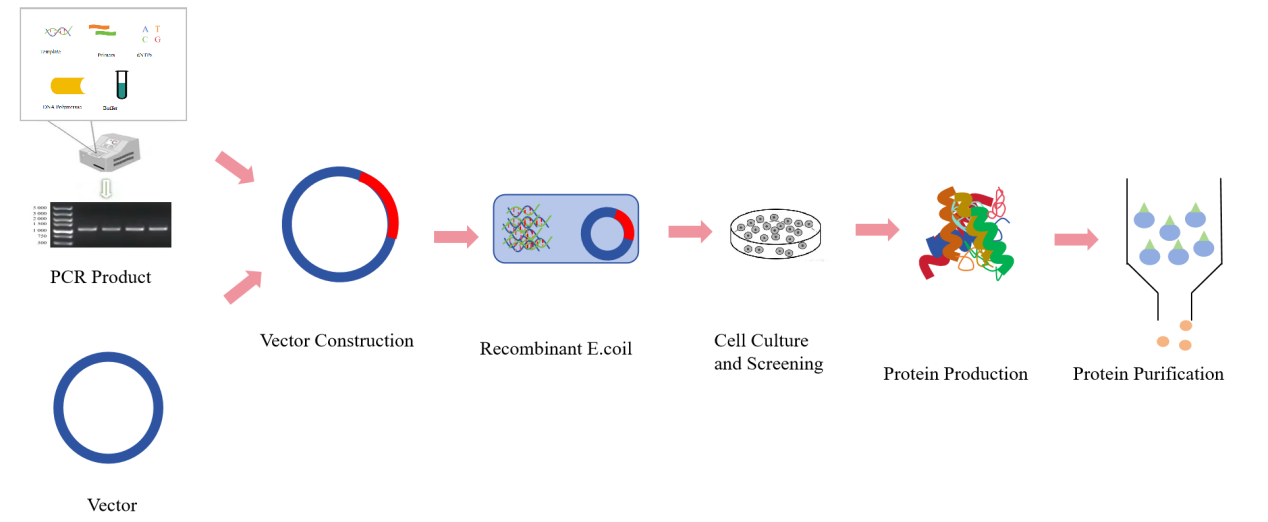
Figure 1:Diagram of prokaryotic expression of recombinant protein
Selection Strategies For Protein Expression Systems
It is important to select the appropriate expression host or use the correct purification method, considering the properties of the target recombinant protein, such as signal peptide, transmembrane binding, solubility, molecular weight and expression location.
(1) Prokaryotic expression system: E. coli is one of the commonly used recombinant protein expression host cells. Its advantages are clear genetic background, rapid reproduction, low cost, high expression, stable 2H/13C/15N isotope labeling technology and wide application range. The disadvantage is that there is no transcriptional and post-translational processing function, which cannot be glycosylated and phosphorylated, and the expressed proteins are easy to form inclusion bodies.
(2) Yeast expression system: Yeast expression system includes saccharomyces cerevisiae and Pichia Pastoris. These yeast cells have the characteristics of eukaryotic cells, which can carry out correct protein folding and modification, carry out processing of expression products, and maintain the activity and stability of products. At the same time, yeast cells can also be large-scale culture and high expression, suitable for the expression of some complex proteins, will not produce toxins, safe and reliable, and the genetic background is clear, the expression system is quite complete, the expression products are secreted into the extracellular, conducive to purification. The disadvantage is that the exogenously expressed protein is easy to be degraded by the self-secreted protein phenol, and the fermentation cycle is relatively long, which is easy to be polluted. Although the product can be glycosylated, the way is mannan type, which is different from that of mammalian cells.
(3) Mammalian cell expression system: Genes expressed in mammalian proteins are consistent with the types and forms of structural glycogroups of natural proteins, and can normally recombine into multiple subunits. In suspension medium or without human serum medium, high density and high culture capacity can be obtained. The disadvantages are that compared with E. coli, the expression level is lower, the time required to obtain high-expression cell lines is long, and the cost of large-scale culture is high.
(4) Insect baculovirus expression system: insect baculovirus expression system has good safety, will not cause harm to human and environmental pollution. Compared with mammalian cell expression systems, the vector construction cycle is shorter. It can accommodate the insertion of large fragments of foreign DNA, is an ideal carrier for expressing large fragments of DNA, can obtain high level expression of recombinant proteins, and has the ability to express multiple genes in the same cell at the same time. The disadvantage is that the recombinant protein is easily degraded in BEVS.
(5) Cell-free protein expression system: cell-free expression can be expressed in vitro without the need for cell division and complex cellular metabolic reactions. It has the advantages of high expression level, easy operation, simplified purification process and flexibility, and is suitable for the preparation of various types of proteins, including transmembrane proteins, toxic proteins, complex proteins, etc., but it also has the disadvantages of limited post-translational modification and folding problems. The choice to use a cell-free protein expression system also needs to be weighed and judged according to the specific research needs and the characteristics of the target protein.
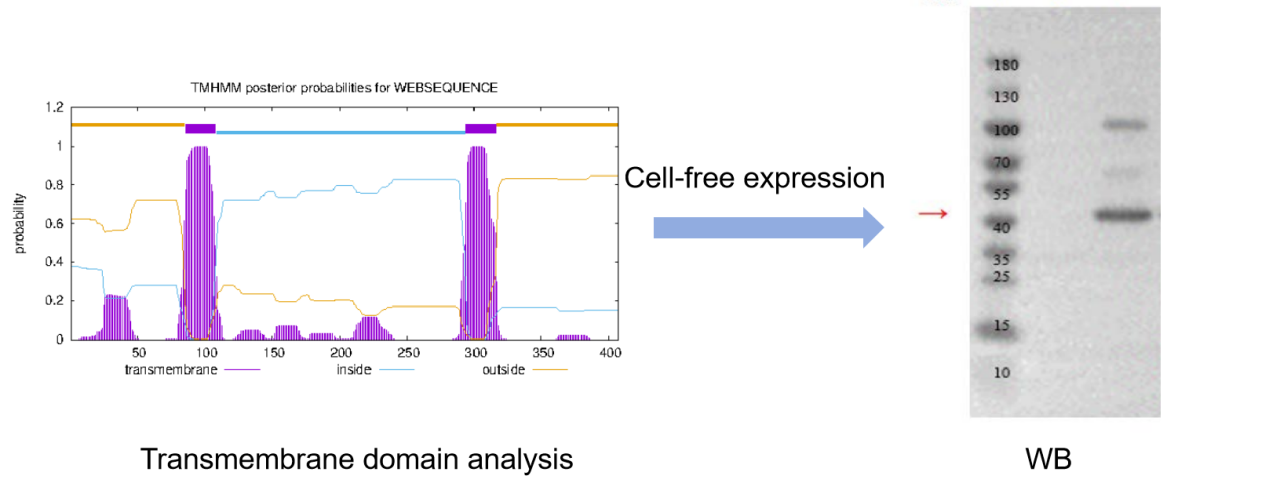
Figure 2:Result of WB validation of cell-free expression of transmembrane protein
Improvement Strategy of Low Protein Expression
(1) Codon optimization, select the most suitable expression vector, and carry out codon optimization during construction, so that the protein can be expressed more completely in the host;
(2) After the plasmid is constructed, it must be confirmed by sequencing, and carefully compare the carrier sequence and insertion sequence to ensure that there is no frameshift and mutation;
(3) Check whether the expression strain is compatible with the plasmid. For example, the pet series vector of Novagen is T7 promoter, which is commonly associated with BL21 and its derivative strains, while the pQE series vector of Qiagen uses T5 promoter, which needs to be expressed by matching strains such as M15.
(4) Check whether the expression conditions are appropriate, such as whether the induced expression is induced by inducer or temperature, and whether it is composed expression;
(5) The selection of medium, LB medium does not express, switching to nutrient-rich TB medium may solve this problem; It is also possible that the protein expression is too low or the degradation is fast, which needs to be confirmed by Western Blot if necessary.
(6) The use of molecular fusion partner technology, the use of promoters, enhancers, tags and other fusion elements to improve protein solubility.
Methods For Quality Determination of Recombinant Protein
(1) SDS-PAGE (Polyacrylamide gel electrophoresis) : SDS-PAGE can separate different proteins in the protein mixture, and the molecular weight of the protein can be determined by comparing molecular weight markers. Protein samples are mixed with SDS and reducing agents, electrophoresis is performed, and then stained with Coomassie brilliant blue or silver staining to analyze protein bands.
(2) Western blot (Western blot) : The use of specific antibodies to detect a specific protein. The proteins separated by SDS-PAGE were transferred to cellulose nitrate or PVDF membrane for immunoassay with specific antibodies.
(3) MS (mass spectrometry) : Identify proteins by determining their molecular weight. The protein is digested into peptides, which are then analyzed using a mass spectrometer.
(4) HPLC (High performance liquid chromatography) : The protein is separated by chromatographic column and its purity and structure are analyzed. Protein samples are injected into HPLC systems for separation and analysis by different types of columns.
(5) ELISA (Enzyme-linked immunosorbent assay) : The presence of the protein to be detected is detected by binding the protein to the specific antibody and using the enzyme labeled secondary antibody. ELISA is suitable for high-throughput sample processing and can also be used to detect the expression levels of a large number of different proteins.
These methods can be used alone or in combination to obtain comprehensive information about the quality, purity, structure, and function of recombinant proteins.
References
[1] Daly R , Hearn M T W .Daly, R. & Hearn, M.T. Expression of heterologous proteins in Pichia pastoris: a useful experimental tool in protein engineering and production. J. Mol. Recognit. 18, 119-138[J].Journal of Molecular Recognition, 2005, 18(2):119-138.DOI:10.1002/jmr.687.
[2] Thomas,J,Magliery,et al.Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism.[J].Journal of the American Chemical Society, 2005.
[3] Fu-Pang,LinKuen-Lin,Leu.Cloning, expression, and characterization of thermostable region of amylopullulanase gene from Thermoanaerobacter ethanolicus 39E[J].Applied Biochemistry & Biotechnology, 2002.DOI:10.1385/ABAB:97:1:33.