2025-04-11 Hits(44)
Protein Interaction
Overview of Protein-protein Interaction
Protein-protein interaction (PPI) refers to the formation of stable or unstable complexes between two or more proteins through the interaction of amino acid residues, such as hydrogen bonds, ionic bonds, hydrophobic interactions, van der Waals forces, and dipole interactions. Typically, the interaction sites of proteins are some specific regions on their surface that have complementary shapes and chemical properties that allow the two proteins to recognize and bind to each other, a process that is specific, and a particular protein usually interacts only with a particular partner protein. Protein interactions can be transient or persistent, and such interactions can change with changes in the cellular environment.
Protein interactions are the basis of biological processes such as intracellular signaling, metabolic regulation, cell cycle control, DNA replication and repair. Protein interactions regulate protein activity, stability, localization, and function, and can mediate signaling inside and outside the cell, for example, receptors on the cell surface bind to signaling molecules and activate a series of protein interactions that transmit signals to the cell interior. It can also be involved in gene expression regulation, for example, after transcription factors bind to DNA, they may need to interact with other proteins to initiate or inhibit gene transcription.
The study of protein interaction is an important field in molecular biology and cell biology, which plays a vital role in understanding the biological phenomena and the mechanism of disease. Understanding protein interactions helps to reveal the complexity and dynamics of biological systems, which is of great significance for the study of disease mechanism and the development of new drugs. However, the intracellular protein interaction network is very complex, involving a large number of proteins and a variety of ways of interaction, so high-precision and high-throughput experimental techniques are needed to identify and resolve the interaction relationships between proteins.
Methods for Analyzing Protein Interaction
(1) Surface plasmon resonance
Surface plasmon resonance (SPR) affinity assay is a biophysical technique used to analyze interactions between biomolecules. It enables real-time monitoring of biomolecular binding events, without labeling, and can provide detailed information on binding affinity, kinetics, and thermodynamic parameters.
SPR technology is based on the phenomenon of surface plasmon resonance, when light strikes a metal surface (usually gold or silver) at a specific Angle, it creates a surface plasmon wave at the metal-medium interface. If the metal surface is bound to biomolecules, the characteristics of the surface plasma wave will change, and this change can be measured by the detection instrument. SPR affinity assay has been widely used in biomedical research and drug development because of its high sensitivity, real-time monitoring ability and no labeling.
.png)
Figure 1:Schematic diagram of surface plasmon resonance technology[2]
(2) Biolayer Interferometry
Biolayer Interferometry (BLI) can detect and analyze biomolecular interactions, based on the principle of light interference, when light passes through two interfaces, interference patterns are generated. In BLI instruments (Fortebio Octet), a sensor tip is usually used with a specific molecule (such as an antibody) attached to its surface. When the target molecule (such as an antigen) binds to the molecule on the sensor tip, it causes the sensor tip surface thickness to change, thus changing the interference pattern. By detecting changes in this pattern, the binding and decoupling processes between molecules can be monitored in real time. BLI technology is widely used in drug discovery, protein engineering, antibody screening and characterization.
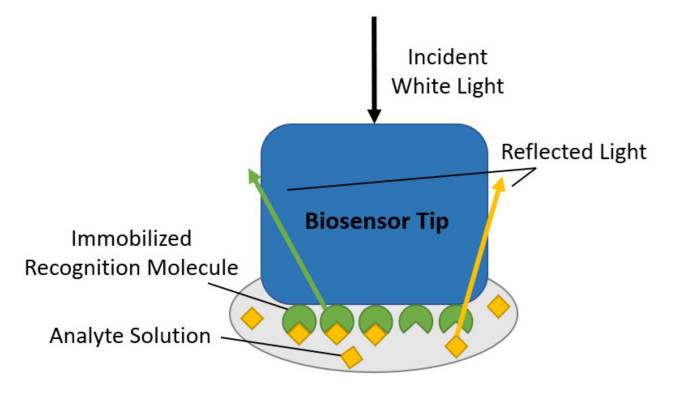
Figure 2:Schematic diagram of Biolayer Interferometry[3]
(3) Co-Immunoprecipitation
Co-Immunoprecipitation (Co-IP) involves the use of antibodies to trap a protein within a protein complex for analysis of the proteins that interact with it. The antibody of the decoy protein is fixed on the solid support, the decoy protein is bound to the antibody, and then incubated with the sample to bind it to the prey protein. After precipitation and washing, the decoy is resuspended in the electrophoresis sample buffer, boiled for 5-10min, the antigen and antibody are dissociated under high temperature and the action of reducing agent, and the supernatant is collected by centrifugation. The supernatant includes antibodies, target proteins and a small amount of miscellaneous proteins. The results of immunocoprecipitation experiment have the advantage of being closer to the protein interaction under real physiological conditions.
(4) Yeast two-hybrid
Yeast two-hybrid, by fusing two proteins with the DNA-binding domain and the activator domain of the yeast transcription factor, respectively, can drive reporter gene expression if the two proteins interact within the yeast cell. The basic principle is that many transcription factors contain two functional domains that are independent of each other, namely the DNA-binding domain (BD) and the activedomain (AD). Transcription factors bind to specific sequences on DNA via BD and AD respectively, thus initiating transcription of the corresponding genes. The experimental procedures included: creating BD-X decoys, testing self-activation by BD-X decoys, performing yeast two-hybrid screening, and identifying prey. The advantage of yeast two-hybrid technology is that it can screen unknown proteins interacting with known proteins in high throughput, but it also has some limitations, such as false positive results and the inability to detect unstable or instantaneous interactions. Therefore, when applying this technique, it is necessary to carefully interpret the experimental results and verify them in combination with other biological methods.
(5) Pull down
Protein pull down experiments utilize specific affinity tags or antibodies to capture the target protein and its interacting partner. This method can be used to determine direct interactions between proteins, or to identify specific protein complexes in cell extracts. The experimental procedures include: selection of affinity labels or antibodies, cell lysis, binding affinity materials, washing, elution, and analysis. By using pull down, we can determine protein interaction network, identify protein complex, and study protein function.
Application of Protein Interaction
The study of protein interaction has a wide range of applications in the field of biology and medicine, which not only enhances our understanding of the basic principles of biology, but also provides new ideas and methods for the diagnosis, treatment and prevention of many diseases. Here are some of the main application areas.
(1) Basic research
Understanding cell signaling: Protein interactions are the basis of cell signaling, and studying these interactions can reveal how signals are transmitted within cells.
Resolving protein function: By identifying partners that interact with a particular protein, it is possible to infer its possible function and mechanism of action.
Construct protein networks: Protein interaction networks help to understand complex interactions within cells and provide data support for systems biology.
(2) Drug discovery
Target identification: Through protein interaction studies, new drug targets can be discovered, which may be key regulators of disease.
Drug design: Understanding the interactions between proteins can help design small molecule drugs to interfere with interactions in pathological states.
Drug retargeting: Known drugs may be used to treat other diseases through novel protein interaction pathways.
(3) Study of disease mechanism
Disease-related protein interactions: The study of protein interaction changes in disease states is helpful to understand the molecular mechanism of disease.
Discovery of disease markers: changes in some protein interactions may be used as biomarkers for disease diagnosis or prognosis.
(4) Gene expression regulation
Transcription factor studies: Protein interactions play an important role in transcriptional regulation, and studying these interactions helps to understand how genes are regulated.
(5) Clinical application
Personalized medicine: Understanding the protein interaction pattern of individual patients can provide a basis for personalized treatment.
Disease treatment: Therapy based on protein interaction, such as the use of antibodies to block specific protein interactions, for the treatment of diseases such as cancer.
FAQS
Q1: High background of target protein.
A: The causes may include non-specific protein binding or non-specific adsorption on the transfer membrane. Solutions include lysis of cells in serum-free cultures, care in laboratory procedures, avoidance of non-specific adsorption, ensuring that all laboratory reagents and equipment have been adequately cleaned and decontaminated, using more stringent washing conditions to reduce non-specific binding, and considering the use of different antibodies or tag proteins to reduce background signals.
Q2: The sample was degraded by protease.
A: It is necessary to add protease inhibitors to the cracked samples and pay attention to the operation on the ice to avoid repeated freeze-thaw. When the antibody concentration is too low, it is necessary to adjust the antibody concentration, and set up a concentration gradient if necessary to find the best concentration.
Q3: Low signal or no signal.
A: Make sure that the antibody or labeled protein used is effective and suitable for the experiment, check that the protein expression and purification procedures are correct, increase the amount of input protein or optimize the experimental conditions.
Q4: Low elution efficiency.
A: Optimize the composition and conditions of the elution buffer and increase the number or time of elution steps.
Q5: Low combination efficiency.
A: Check the expression level and purity of the fusion protein, and optimize the conditions of the binding reaction, such as time, temperature, and ionic strength.
References
[1] Hakes L , Pinney J W , Robertson D L ,et al.Protein-protein interaction networks and biology--what's the connection?[J].Nature Biotechnology, 2008.DOI:10.1038/nbt0108-69.
[2] Sultana A , Lee J E .Measuring Protein‐Protein and Protein‐Nucleic Acid Interactions by Biolayer Interferometry[J].Current protocols in protein science, 2015, 79:19.25.1.DOI:10.1002/0471140864.ps1925s79.
[3] Murali S, Rustandi RR, Zheng X, Payne A, Shang L. Applications of Surface Plasmon Resonance and Biolayer Interferometry for Virus-Ligand Binding. Viruses. 2022 Mar 29;14(4):717. doi: 10.3390/v14040717. PMID: 35458446; PMCID: PMC9027846.