2025-01-10 Hits(125)
Peptide Library
The peptide display library consists of many peptides with specific lengths and different sequences, including most of the combinations of amino acid sequences. According to the source, it can be divided into synthetic peptide library, native peptide library, and recombinant protein peptide library. The synthetic peptide library comprises various peptide sequences prepared by chemical synthesis. The native peptide library is composed of natural peptides in living organisms. The recombinant protein peptide display library was constructed based on protein engineering technology. It is used in drug design, protein-protein interactions, and other biochemical and drug research fields.
The current methods of peptide display library screening and construction are as follows:
Phage display has the advantages of a mature research system, low cost, and operating room list. Using this technology to develop cyclic peptide drugs can provide a new strategy for the screening of cyclic peptide drugs. Phage peptide library synthesis and screening are as follows:
(1) Principle
The construction of the phage display cyclic peptide library is to display peptide segments and proteins on pⅢ or pVⅢ protein of M13 filamentous phage, which is suitable for screening high-affinity cyclic peptide ligands for specific targets. Dicyclic peptides are more complex and stable and have been developed to treat tumors. Dicyclic peptide-drug conjugate has been generated and is being evaluated in clinical trials.
(2) Screening method
Solid phase peptide synthesis involves fixing the target protein directly (via covalent bonds) or indirectly (using biotin or streptavidin splices) to a solid support, incubating it with a phage library for some time, and washing it multiple times. The process was repeated 3-4 times to obtain the phage display library. The liquid phase peptide synthesis is screened for the biotin-labeled target protein by incubation, and then the bound phage is captured using magnetic beads. Solid phase peptide synthesis consumes more proteins, and the natural epitopes of proteins will be destroyed. The liquid phase peptide synthesis has high efficiency and enrichment. The way of phage library screening depends on the specific experimental requirements, which will have a certain impact on the effectiveness of the phage cyclic peptide library.
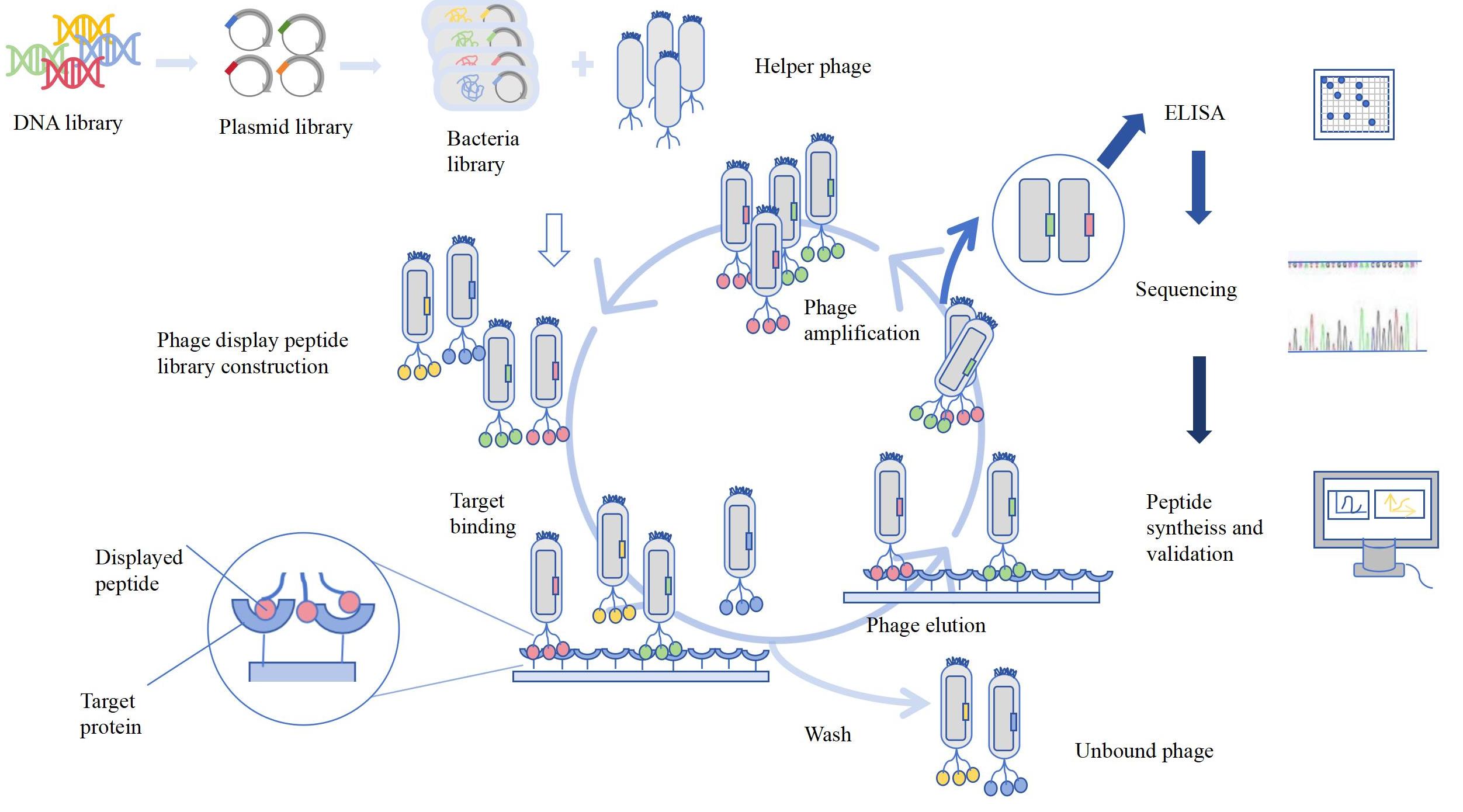
Fig 1: Phage display peptide library construction and screening process.
2. mRNA display technology
The principle of mRNA display technology is to transcribe the DNA library or translate the mRNA library in vitro to form a cyclic polypeptide, then generate an mRNA-cDNA double strand through reverse transcription to prepare the MRNA-cyclic peptide library. The mRNA-cDNA was incubated with a specific protein, and the unbound polypeptide was removed by washing several times. PCR was used to amplify the cDNA encoding the target protein-binding polypeptide as a template to enrich the target protein-binding polypeptide.
3. One-bead-one-compound technology
The technique can synthesize a large number of different compounds in a short time and can help researchers quickly find compounds with specific properties. It is used in research of polypeptides and oligonucleotides, proteins, synthetic oligomers, small molecules, and oligosaccharides.
The screening methods of the one-bead-one-compound polypeptide library are as follows:
(1) Magnetic separation method is to incubate the bead-to-object polypeptide library with the biotinizing target and streptavidin-coated magnetic particles. The positive polypeptide beads were combined with magnetic particles to make the polypeptide beads magnetic. At this time, placing a magnet outside the container can adsorb the positive polypeptide beads to the container wall, the negative polypeptide beads will sink to the bottom of the container. The disadvantage of magnetic sorting is that it has a high false positive rate and usually requires multiple screening.
(2) Enzyme-linked immunosorbent refers to antigenic antibodies of ligands (usually proteins) by specific binding methods. In the enzyme-related immunosorbent process, a bead-to-object polypeptide library is incubated with the routinized target, and then streptavidin alkaline phosphatase conjugate is added.
(3) The binding of the ionized target to the polypeptide beads enriched streptavidin alkaline phosphatase to the surface of the polypeptide beads, resulting in turquoise precipitates on the positive polypeptide beads. The darkest polypeptide beads were manually separated using a micropipette device and thoroughly cleaned to remove turquoise fluorescent probes and remaining proteins. Fluorescence-based screening of a fluorescent probe to a specific protein and then co-incubate it with a bead-to-object polypeptide library. Under certain conditions, some fluorescent probes emit strong fluorescence to light up the beads, then manually screen positive polypeptide beads. The disadvantage of fluorescence-based screening is that it is slow and requires manual operation by the operator under an optical or fluorescence microscope.
KMD Bioscience has rich experience in antibody engineering construction and mature antibody humanization technology that can provide customers with peptide library synthesis and screening techniques, phage display technology, mRNA display technology, and other services. In addition, KMD Bioscience has many years of experience in phage cyclic peptide library screening, so we can also provide polypeptide library screening services, phage library screening services, and antibody customization services, including antibody expression and purification, affinity determination, antibody sequencing, etc., to meet customer needs.
References:
[1] Sohrabi C, Foster A, Tavassoli A. Methods for generating and screening libraries of genetically encoded cyclic peptides in drug discovery [J]. Nat Rev Chem. 2020, 4(2): 90-101.
[2] Ji X, Nielsen AL, Heinis C. Cyclic Peptides for Drug Development [published correction appears in Angew Chem Int Ed Engl [J]. 2024, 63(10): e202319807.
[3] Hu LY, Kelly KA, Sutcliffe JL. High-Throughput Approaches to the Development of Molecular Imaging Agents [J]. Mol Imaging Biol. 2017, 19(2): 163-182.