2024-12-18 Hits(101)
Phage Display
As a company focused on antibody development, KMD Bioscience provides one-stop services from immunogenic synthesis, animal immunization, Fab antibody library construction and screening, identification and optimization, with HEK293 or CHO cells as the expression host, and ultimately the delivery of high-quality recombinant Fab antibody fragments.
Antibody and Fab Antibody
Antibody Characterization: Antibody, also known as immunoglobulin, is a kind of immunoglobulin that is synthesized and secreted by the body's immune cells after they are activated by the antigen and the B cells differentiate and mature into plasma cells. The basic structure of Ig molecule is composed of four polypeptide chains. That is, it consists of two identical peptide chains with relatively large molecular weights (called heavy chains) and two identical peptide chains with relatively small molecular weights (called light chains). The light chain and the heavy chain are connected by disulfide bond to form a tetrapeptide chain molecular monomer called Ig molecule, which is the basic structure of immunoglobulin molecule.
Fab antibody characterization: Fab antibody characterization is multifaceted, including antigen-binding properties, molecular weight size, stability and others. Fab antibody is an antigen-binding fragment, which is the antigen-binding region of an antibody. The Fab fragment can be further divided into two parts: Fv fragment composed of light chain variable region and heavy chain variable region, and Fb fragment composed of light chain constant region CL and heavy chain constant region CH1. There is an antigen-binding site between each heavy and light chain, which allows Fab antibodies to bind to multiple epitopes on the surface of the antigen, thereby enhancing its affinity and specificity. The Fab fragment contains 440~450 amino acids and its molecular weight is about 55kDa.
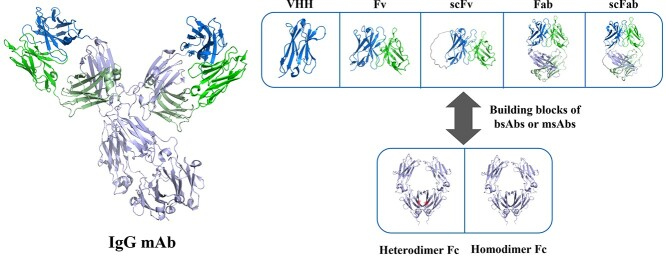
Fig.1 Structural diagrams of IgG monoclonal antibodies (IgG mAb) and Fab antibodies. (From “Developability assessment at early-stage discovery to enable development of antibody-derived therapeutics.”)
Compared with complete antibodies, Fab antibodies have many unique advantages, such as no Fc fragments, low relative molecular mass, high fluidity and tissue penetration, short cycle half-life, high binding force, and low immunogenicity. The development of Fab antibodies has undergone a transition from natural antibodies to genetically engineered antibodies. On the basis of genetically engineered antibody, Fab antibody as an important antibody fragment has gradually entered people's field of vision.
Fab Antibody Production Method
Enzymolysis: Fab fragments can be obtained by enzymolysis of complete IgG antibodies with enzymes such as papain or pepsin. This method is quick and easy, but may lose some immune reactivity.
Genetic engineering: Using genetic engineering methods such as phage display technology or mammalian cell expression systems, engineered Fab antibodies can be produced. These antibodies have higher affinity and specificity, and can be modified and optimized as needed.
Antibody Developability Assessment
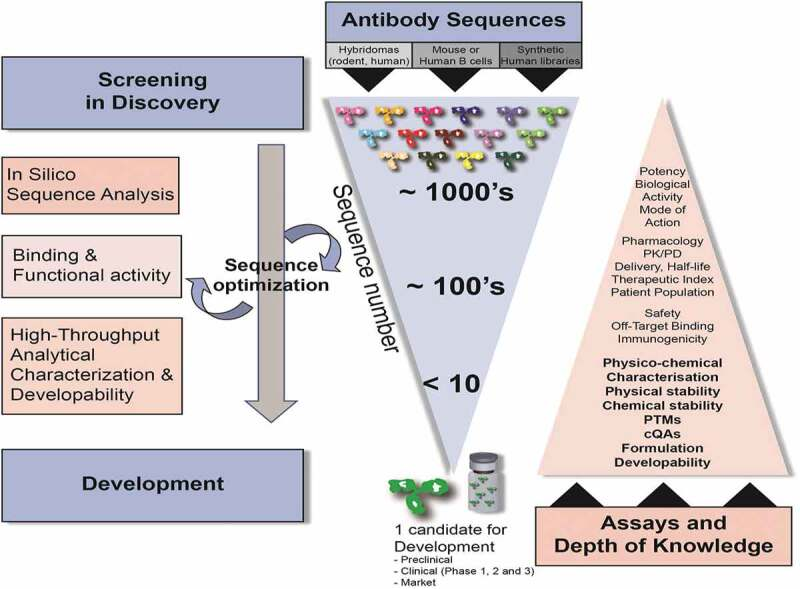
Fig.2 Medicine discovery, sequence selection, and exploitable workflows. (From “Predicting Antibody Developability Profiles Through Early Stage Discovery Screening.” )
Monoclonal antibodies are playing an increasingly important role in the development of new medicines in multiple therapeutic areas. During early antibody generation and screening, antibody developability assessment through implementing efficient and practical high-throughput exploitable workflows is critical to selecting the best lead medicine candidates.
Antibody developability assessment covers the feasibility of further analyzing a molecule's progress from discovery to successful development by evaluating its physicochemical properties, including the likelihood that antibody medicine candidates will become producible, safe, and effective medicines. These properties include the tendency of self-interaction and aggregation, thermal stability, colloidal stability, etc. The optimal antibody molecule is selected based on biological function, efficacy, safety and developability.
To reduce the risk of poorly developable antibody medicine candidates entering the CMC stage, candidates should be screened, evaluated, and optimized for development-related properties as early as possible. Antibody developability assessment in the early discovery phase should be conducted in a rapid, high-throughput manner while consuming a small amount of test material.
The application field of antibody development technology is very wide, including viral infection, tumor treatment, autoimmune disease and many other aspects. KMD Bioscience’s antibody development platform utilizes modern biotechnology methods, such as phage display technology, single B cell technology, hybridoma technology, etc., to synthesize and prepare these antibodies with specific functions for customers.
Techniques of Antibody Development
By fusing antibody-producing B lymphocytes with myeloma cells, hybridoma cells are formed. These cells can both produce antibodies and proliferate indefinitely, producing large amounts of antibodies.
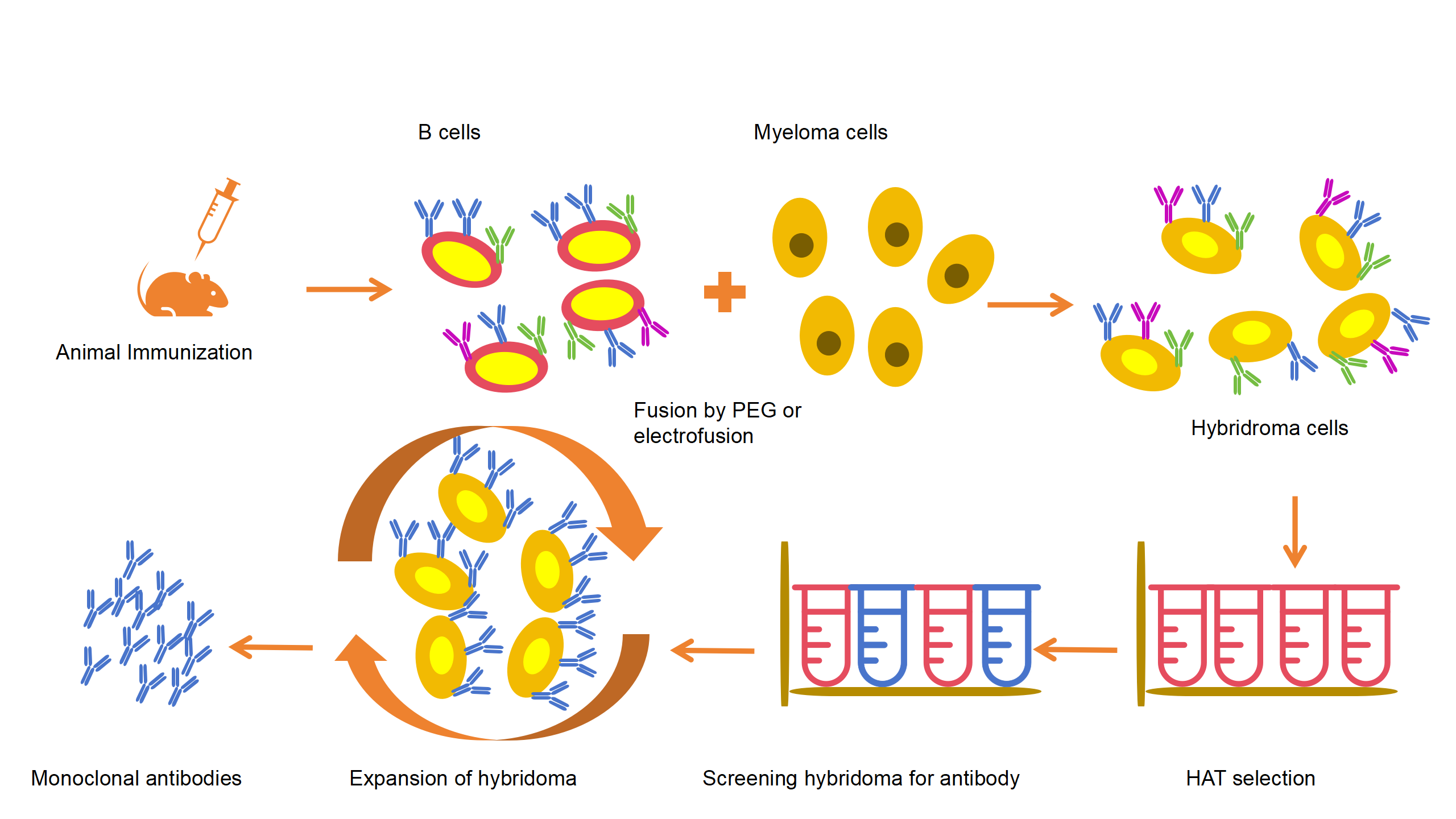
Fig 3: The process of producing antibodies using hybridoma technology.
By isolating a single B cell, extracting its antibody gene, and using genetic engineering technology to express, so as to prepare the corresponding antibody.
.png)
Fig 4: The process of producing antibodies using single B cell technology.
The DNA that codes for foreign peptides or proteins is inserted into the phage's genome so that these peptides or proteins are displayed on the phage's surface. Through screening, antibodies with high affinity to specific targets can be obtained.
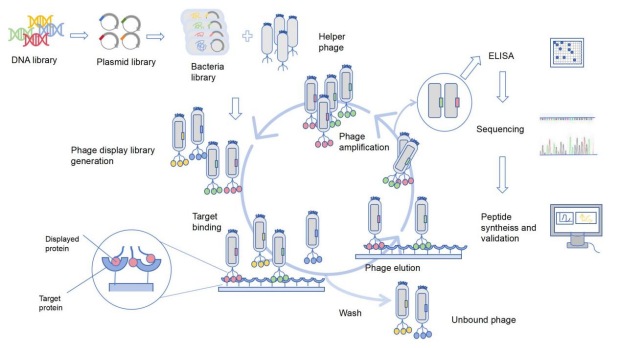
Fig 5: The process of producing antibodies using phage display technology.
Antibody Optimization
The main goal of antibody optimization
Improve affinity: Through the introduction of mutations and screening, enhance the ability of the antibody to bind to the target, thereby improving the efficacy of the antibody.
Improve stability: including physical stability and chemical stability, to avoid degradation and inactivation of antibodies during storage and use.
Improve specificity: Reduce the cross-reaction of antibodies with non-target, improve the accuracy of treatment and diagnosis.
Reduce immunogenicity: Make the antibody closer to the structure of the human autoantibody, reducing the immune response in the human body.
The main methods of antibody optimization
Directed evolution and random mutation
Directed evolution: By simulating the process of natural selection, antibodies are screened and optimized for multiple rounds to screen out antibody variants with higher affinity and stability.
Random mutation: Random mutations are introduced into antibody genes using mutation techniques (such as PCR-mediated mutation), and then antibody mutants with excellent properties are screened by high-throughput screening techniques.
Protein engineering technology
Genetic engineering methods are used to modify antibodies, such as humanization (transforming murine antibodies into human antibodies to reduce immunogenicity) and antibody fragmentation (cutting antibodies into smaller fragments to improve penetration).
In vitro evolutionary technique
Such as phage display technology, yeast display technology and cell surface display technology, these technologies can screen and optimize antibodies in vitro environment, so as to quickly obtain high affinity and high stability of antibodies.
Reference
[1]. Zhang W, Wang H, Feng N, Li Y, Gu J, Wang Z. Developability assessment at early-stage discovery to enable development of antibody-derived therapeutics. Antib Ther. 2022 Nov 11;6(1):13-29.
[2]. Bailly M, Mieczkowski C, Juan V, et al. Predicting Antibody Developability Profiles Through Early Stage Discovery Screening. MAbs. 2020;12(1):1743053.