Service Line:+86-022-82164980
Address:FL-4, Building A5, International Enterprise Community, Tianjin, China
Email:[email protected]
KMD Bioscience has been dedicated to protein expression and protein sequencing research for over 10 years. Amino acid sequence analysis of proteins is the foundation and key to protein research. The information obtained through protein sequencing has many valuable applications, such as protein identification, molecular cloning probe design, and immunogen peptide synthesis. Based on high-resolution mass spectrometry, We can provide customers with high-fidelity antibody and protein sequencing services.
KMD Bioscience has highly skilled laboratory technicians with extensive experience in protein and antibody sequencing and can provide mass spectrometry-based protein sequencing analysis services. Combined with bioinformatics databases, our protein sequencing platform can realize rapid and accurate protein sequence analysis of protein primary structure, and can also provide N-terminal protein sequencing services.
KMD Bioscience is equipped with a high-quality Orbitrap Fusion Lumos mass spectrometer and has established a perfect database, which can provide high-quality protein sequencing service for our customers. Given different experimental needs, we also provide researchers with the Edman degradation method to determine the length of up to 67 amino acid sequences at the N-terminus of proteins.
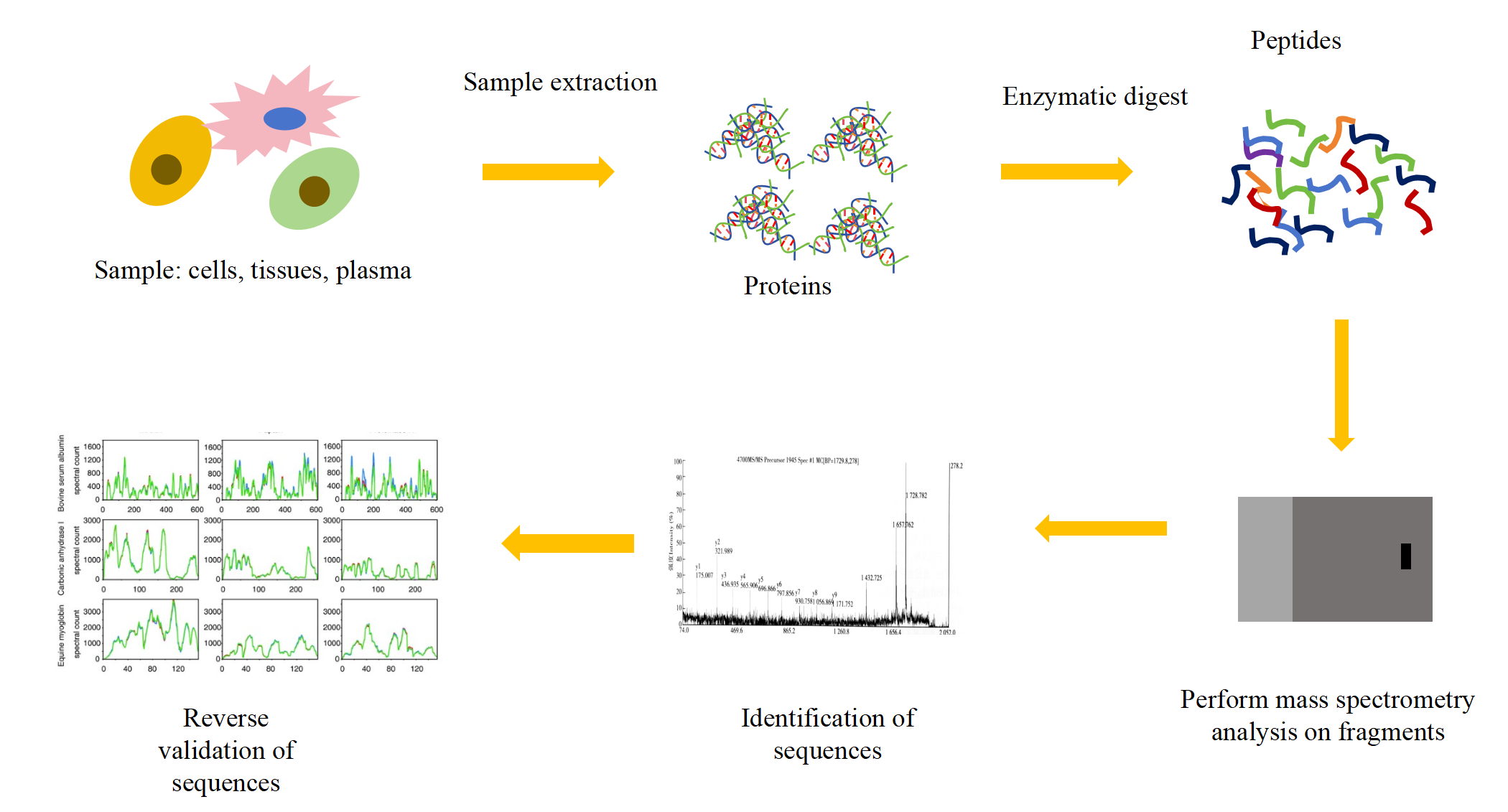
Generally, the protein is digested into a peptide of 5-25 amino acids by mass spectrometry, then analyzed and detected by a mass spectrometer, and the collected data is matched with the theoretical sequence database. Mass spectrometry can also perform de novo protein sequencing, providing a solution for the determination of a completely new or no theoretical database of protein sequences.
After the customer sends the protein product to KMD Bioscience, we will first identify and fragment the protein sample using the commonly used 6 proteases, including Trypsin, Chymotrypsin, Asp-N, Glu-C, Lys-C, and Lys-, to get as many kinds of fragmented sequences as possible and realize the full coverage of protein sequences. Sequence information from peptide mass spectrometry sequencing is spliced by PEAKs Studio (for protein products) and PEAKs Ab (for antibody products) software with manual assistance to get correctly spliced protein or antibody primary sequences.

Figure 1 Orbitrap Fusion Lumos Mass Spectrometer
--KMD Bioscience sequencing service implements strict quality control and adopts the principle of wide-in, narrow-out, which realizes the accurate identification of protein sequences without omitting valid data information.
--Compared with the previous generation Orbitrap Elite mass spectrometer, the Orbitrap Fusion Lumos mass spectrometer used in our laboratory improves the speed of primary and secondary mass spectrometry scanning while ensuring the accuracy of peptide molecular weight.
--Combined with our sequence splicing logic algorithm and the processing power of our commercial software (based on PEAKS software analysis), we can differentiate amino acids with close molecular weights. For example, we can achieve differential analysis of glutamine and lysine with a molecular weight difference of 0.03Da.
--Our sequencing platform can distinguish different variants of homologous amino acids, and the characteristic peaks of secondary fragmentation fragments can accurately determine leucine and isoleucine. Our system is also capable of distinguishing amino acid molecules with post-transcriptional modifications.
--Our protein sequencing platform not only includes de novo protein sequencing but also provides other sequencing services such as N-terminal sequencing, C-terminal sequencing, Edman sequencing, de novo peptide sequencing, de novo genome sequencing, etc.
--Protein samples: >90% purity (SDS-PAGE result), at least 100ug
--Try not to contain BSA in the sample. Prior communication is required before sending samples
If you have any questions regarding our services or products, please feel free to contact us by E-mail: [email protected] or Tel: +86-400-621-6806