Service Line:+86-022-82164980
Address:FL-4, Building A5, International Enterprise Community, Tianjin, China
Email:[email protected]
KMD Bioscience has been committed to innovative drug discovery services for many years and has rich experience in aptamer development (including aptamer development for proteins, peptides, amino acids, small molecule substances, metal ions, etc.). By flexibly utilizing the natural evolution of ligand systems through exponential enrichment (selex screening) method (such as SELEX method can identify different cells or detect molecular level differences between two types of cells), KMD Bioscience provides in vitro aptamer screening services. Customers only need to provide the sequence number of the target molecule, and KMD Bioscience can screen the aptamer sequence (RNA aptamer or DNA aptamer sequence) for the target molecule quickly, efficiently, and accurately. At the same time, KMD Bioscience can also provide customers with one-stop services from gene analysis synthesis, in vitro screening of aptamers, aptamer synthesis, to affinity determination, helping different customers conduct experiments and providing strong support for downstream research and development work such as affinity verification, competitive ELISA verification, in vitro targeted cell functional verification (such as in vitro recognition and inhibition function verification of nucleic acid aptamers, in vitro flow cytometry blockade function, etc.), in vivo functional verification (such as in vivo targeted inhibition function verification of aptamers, signal pathway blockade function verification, etc.), and targeted specific molecular drug development.
KMD Bioscience can provide adapter selex screening services for various samples (including proteins, peptides, amino acids, small molecules, etc.), and there are also various methods for adapter selex screening (magnetic bead SELEX, cell SELEX, capture SELEX, etc.), among which magnetic bead screening is a commonly used screening method. For some special samples, such as drug small molecules that require screening for aptamers, small molecule modification techniques are often needed to modify them. KMD Bioscience's library has a capacity of 10 ^ 13-10 ^ 14, which is sufficient to screen for nucleic acid aptamers targeting customer targets. After 6-10 rounds of pressure screening, highly specific nucleic acid aptamers can be obtained. Based on the mature SELEX technology platform, the selected DNA aptamers and RNA aptamers can have higher binding affinity than conventional aptamers, reaching the level of nM pM. Scientists at KMD Bioscience will evaluate and design the best solution for small molecule modification and adapter screening based on the client's project requirements.
Small Molecule Nucleic Acid Aptamer Screening Service
Nucleic acid aptamer screening is mainly based on SELEX technology, which selects nucleic acid molecules that highly specifically bind to specific targets from a large nucleic acid library containing different sequences through multiple rounds of selection and amplification processes. The nucleic acid aptamer (DNA aptamer or RNA aptamer) obtained through in vitro screening technology can recognize the corresponding target molecule. The three-dimensional structure of the aptamer determines its high specificity, and therefore can bind specifically to the target molecule. Compared with traditional antibodies, nucleic acid aptamers have the advantages of high thermal stability, chemical synthesis and modification ability, and low immunogenicity. Therefore, nucleic acid aptamers have broad application prospects in many fields such as biological analysis, biomedicine, and sensing technology. Nucleic acid aptamers can specifically bind to target molecules for early diagnosis and screening of diseases. As a targeting molecule, nucleic acid aptamers can accurately deliver drugs to the lesion site, improve therapeutic efficacy, and reduce side effects. Biosensors combined with nucleic acid aptamers can detect biomarkers and environmental pollutants with high sensitivity. With the continuous deepening of research and advances in technology, nucleic acid aptamers will play an important role in more fields. Aptamers can specifically bind to predetermined targets, and they are also defined as "artificial antibodies". The basic idea of SELEX technology is to chemically synthesize a single stranded oligonucleotide library in vitro, which is mixed with the target substance. Through repeated screening and amplification, nucleic acid molecules that do not bind to the target substance are washed away, and nucleic acid aptamers with high affinity for the target substance are isolated and purified. SELEX technology can be used to screen nucleic acid aptamers with high affinity for specific and target substances from a random single stranded nucleic acid sequence library. After Tuerk and his colleagues initially employed this technology to identify specific oligonucleotide ligands capable of selectively binding to bacteriophage T4 DNA polymerase and organic dye molecules, SELEX technology has evolved into a significant research instrument over the span of more than a decade. Over this period, SELEX has offered a precise and efficient approach for screening nucleic acid aptamers, thereby advancing their application and development across diverse domains.
Customers only need to provide the sequence number of the target molecule or small molecule raw material (small molecule substances need to have good water solubility>1mM; stable, not easily oxidized, not easily decomposed, not easily diluted with high heat; sample purity>95%; sample size not less than 10mg), and KMD Bioscience will modify the small molecule for screening nucleic acid aptamers. Through multiple rounds of screening, the abundance of nucleic acid aptamers that can bind to the target molecule gradually increases. Finally, after NGS sequencing, the nucleic acid aptamer sequence specific to the target molecule is obtained. The specific process is shown in Figure 1.
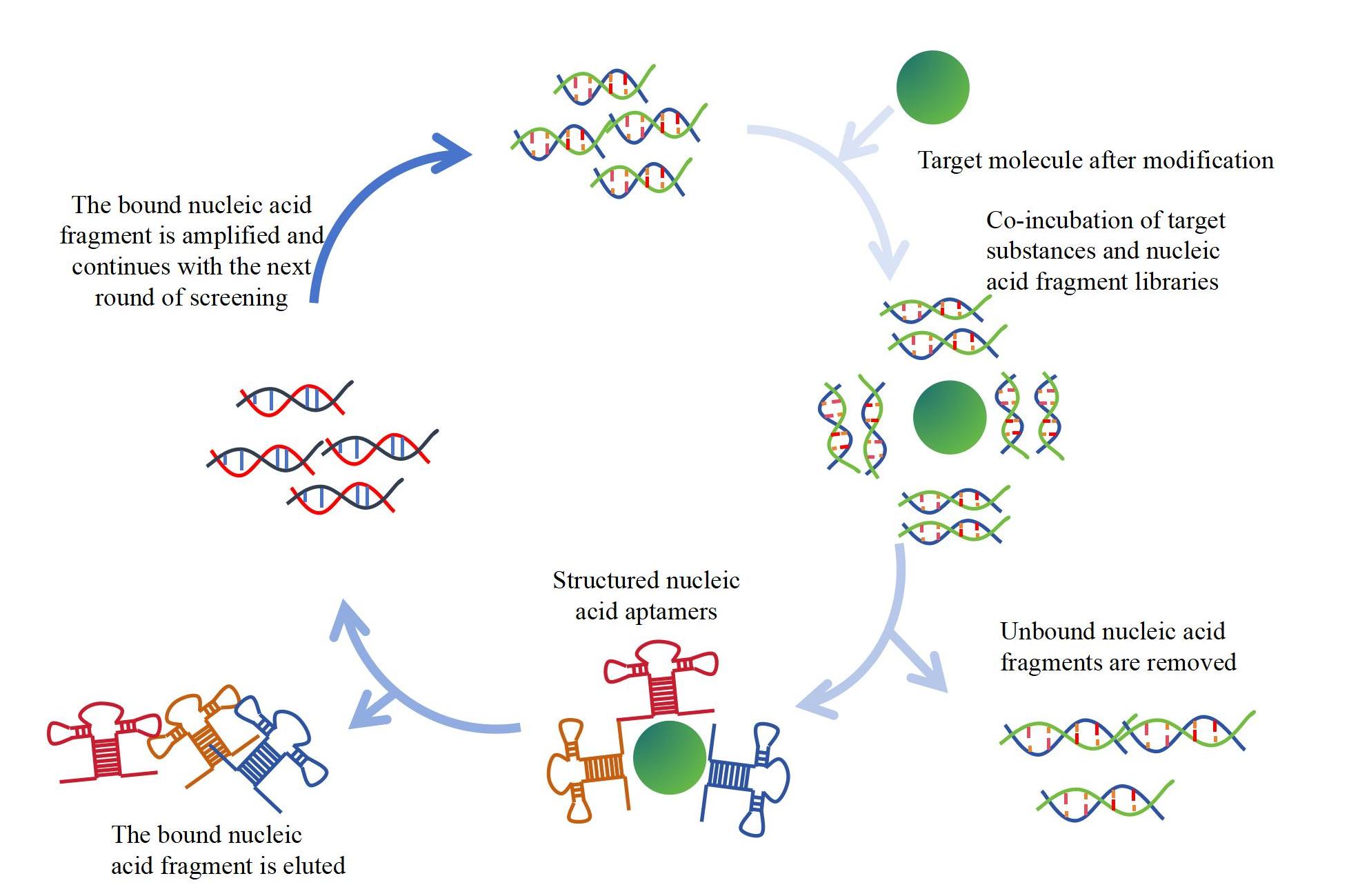
Figure 1 Service process for small molecule nucleic acid aptamer development based on SELEX technology
Small Molecule Nucleic Acid Aptamer Screening Service Workflow
|
Step |
Service Content |
Timeline |
|
Step1:Small molecule modification |
(1) Small molecule raw materials or CAS numbers provided by customers. (2) Small molecule modification: KMD Bioscience couples small molecule biotin. (3) Delivery: 5-10 mg modified product (purity>90%), experimental report, HPLC and MS identification results. |
4-5 weeks |
|
Step 2: Screening of nucleic acid aptamers |
(1) Modified small molecules as screening targets. (2) Adaptation library screening and enrichment: PCR amplification enrichment+transcription+gel running recovery, usually 6-10 rounds. (3) Screening products for NGS sequencing. (4) Delivery: 5-15 adapter sequences, experimental report, raw data (including NGS sequencing raw data and gel electrophoresis) |
10-15 weeks |
|
Step3: Adaptation synthesis and affinity determination (optional) |
(1) The adapter is synthesized according to the sequence. (2) Affinity determination of adapter and target protein, KD determination by BLI or SPR. (3) Delivery: Experimental report, raw data |
4-5 weeks |
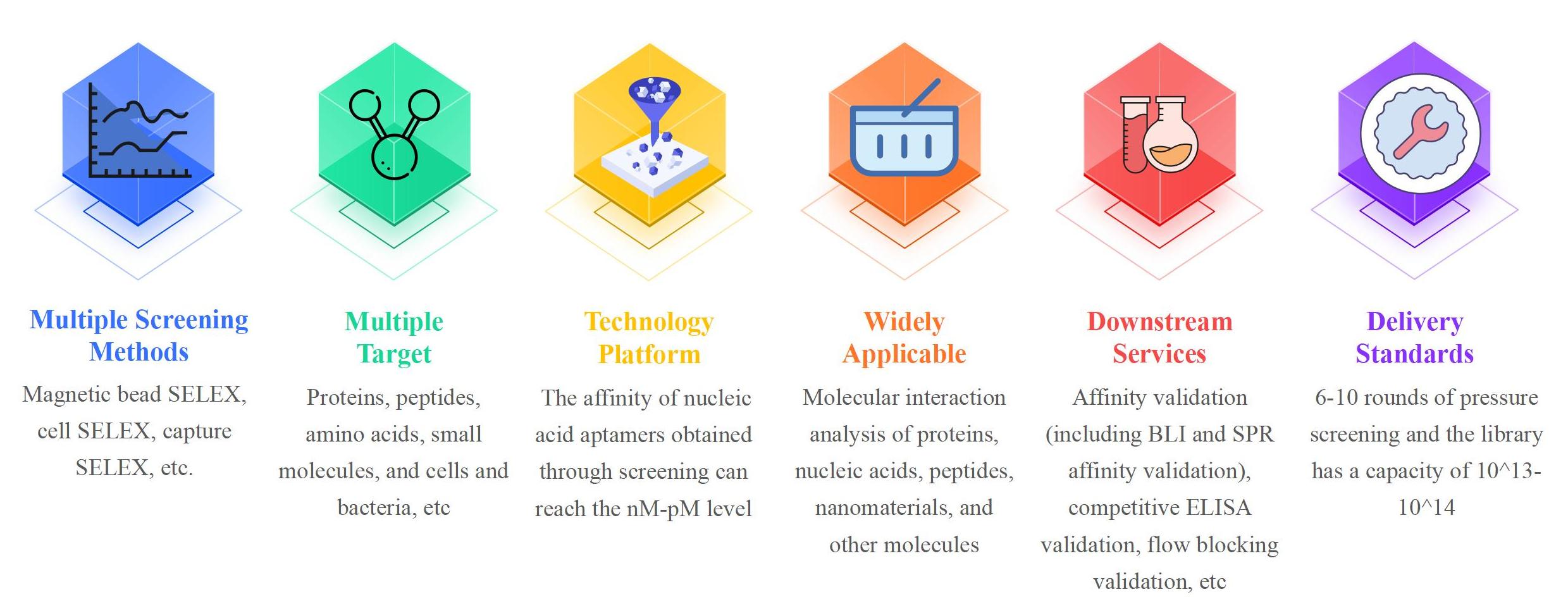
Small Molecule Nucleic Acid Aptamer Screening Service Advantages
FAQ-Small Molecule Nucleic Acid Aptamer
1. What is a small molecule nucleic aptamer?
A: Aptamers, also known as nucleic acid aptamers, are an in vitro screening technique that uses systematic evolution of ligands by exponential enrichment (SELEX) to obtain structured oligonucleotide sequences. The sequences obtained by technical screening can be nucleic acid or deoxynucleic acid. These sequences can recognize them and bind to specific small molecules with strong affinity. Single-stranded oligonucleotides undergo a complex adaptive folding process, forming specific three-dimensional spatial structures using various intermolecular forces such as complementary pairing between nucleotides, hydrogen bond formation, π - π stacking effects, and electrostatic interactions. Next, these three-dimensional structures bind to the target small molecules using intermolecular interaction forces. In recent years, small molecule nucleic aptamer has shown its application potential in the detection of pollutants in the two disciplines of environmental engineering and food science. Small-molecule pollutants are the main detection objects in these two major disciplines. Pollutants include toxins, antibiotics, endocrine disruptors, pesticides, and other substances, which have a great impact on human health and food safety, so how to detect them is now a hot area of research. Due to the importance of contaminants, aptamer screening techniques for small molecule targets have now become a new field for scientists to explore.KMD Bioscience can provide customers with small molecule aptamer services, facilitating the establishment of follow-up testing methods and other applications.
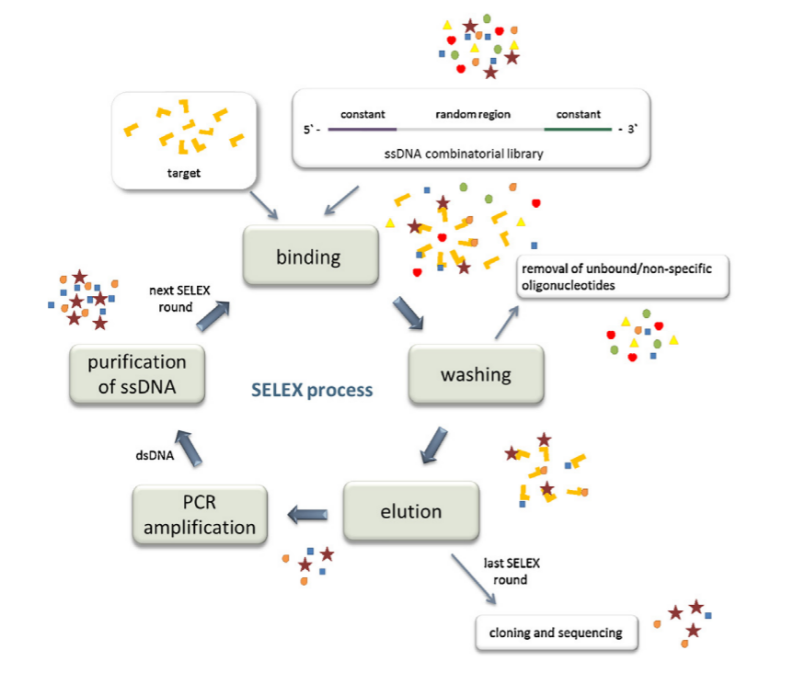
Figure 2: SELEX technology. (Reference documentation:Šmuc T, Ahn IY, Ulrich H. Nucleic acid aptamers as high-affinity ligands in biotechnology and biosensors. J Pharm Biomed Anal. 2013 Jul-Aug;81-82:210-7. )
2. The advantages of small-molecule nucleic aptamers compared with antibodies?
A: As a detection tool in the field of modern scientific research, small molecule nucleic aptamer is considered an alternative product of antibodies and can play its role in the field of detection technology, drug development, and biology. In contrast to the tedious preparation of antibodies, the preparation of aptamer abandoned the reaction and binding in the organism and chose to select the sequence of the oligonucleotide with high affinity capable of binding to the target small molecules in vitro. During the in vitro screening process, the experimenter can adjust various parameters, such as the temperature of incubation, PH, and incubation time, to improve the performance of the best-obtained aptamer. At the same time, after the target sequence is screened and identified, we can use the machine to synthesize the sequence. A high degree of automation in this process can ensure that the differences between the sequences produced in each synthesis are very small. In extreme environments such as high temperature, and strong acid and base environments, the stability of the aptamers is also stronger than the naturally produced antibodies, and they can quickly recover their natural structure, and again bind to the target small molecules. Aptamers are also able to quickly complete labeling, whether fluorescent, nanoparticles, or other functional groups, providing an ideal component choice for biosensor construction.
3. Three kinds of methods commonly used for screening aptamer, and how to solve them?
A: Currently, the three main screening techniques used by scientists are target fixation-based screening technology, innovative Capture-SELEX (library fixation screening technology), and GO-SELEX (SELEX using graphene oxide). Although the method of target fixation may affect the result due to steric hindrance and other conditions, this method is simple to operate and is deeply loved by people. At the same time, the Capture-SELEX technology, which has been widely used in recent years, also needs our special attention. Given the above three technologies, We selected the key experimental conditions such as library design, positive screen target concentration setting, negative screen target selection, and concentration control, and then found the relationship between the affinity and specificity of the aptamer. By analyzing the screening process and results of more than 30 different target aptamers, some scientists found that the affinity of the aptamer can be improved if the concentration of the positive screening target is lowered, but this step is not necessary in the experimental process. Scientists have also found that the specificity of the aptamers obtained by existing technologies makes it difficult to meet some application criteria. We can adopt a negative screening technology that can help us improve the specificity of the aptamer and optimize our screening process.
4. How do small-molecule nucleic aptamers change their performance through various modifications?
A: During the development of small-molecule aptamers, the researchers have discovered and used different modifying groups, including DNA / RNA bases, sugars, and phosphate components, to modify the aptamers. Notable is that the sugar and phosphate group of aptamer modification, helps aptamer improve the ability of nuclease to its degradation, the group after the modification of aptamer ability, can help small molecule aptamer better should be in drug discovery, the development of biosensor and other fields. In addition, some groups can modify the aptamer, which can help stabilize the three-dimensional spatial conformation of the active aptamer, which in turn promotes tight binding to the target small molecules. In addition, some groups can modify the aptamer, which can help stabilize the three-dimensional spatial conformation of the active aptamer, which in turn promotes tight binding to the target small molecules. In the field of small molecule aptamer recognition, the literature enhancing DNA / RNA aptamer specificity by base modification is now scarce. In published studies, researchers have used thymine to replace conventional uracil and added other modified DNA aptamers to maintain binding activity to ATP, despite a reduced affinity. This study demonstrates that the introduction of external functional groups within a reasonable range can expand the application of aptamers without significantly interfering with the aptamer core function.
5. In small molecule aptamers, traditional SELEX screening is labor-intensive and usually has a low success rate. How do we solve this problem?
A: Some scholars have proposed and implemented a new strategy, to study and develop high-affinity aptamers that can efficiently bind to small molecules. The core of this strategy is the use of existing DNA aptamers with thermal stability to give them new functions and targets. First, we collected the DNA aptamer library with excellent thermal stability and then predicted these aptamers using biological techniques. Through this prediction process, we could successfully select alternative aptamers with high affinity. To further test the above predictions, we used MST experiments to measure the strength of the aptamers and small molecules when binding, while we also simulated the interaction mechanism between them. The results of the simulations can also help us to determine whether that base pair plays a key role. In addition, the scientists have explored some non-SELEX screening methods, such as non-equilibrium capillary electrophoresis (NECEEM) of equilibrium mixtures, and non-SELEX capillary electrophoresis technology, which can efficiently express and select nucleic acid aptamers without multiple rounds of screening.Pro-SELEX technology, developed by Northwestern University after years of research, can realize the quantitative screening of aptamers with high binding affinity through microfluidic chips. This method can improve the accuracy and success rate of aptamer screening. KMD Bioscience is committed to providing customers with a short screening cycle, and high successful efficiency of small molecule nucleic acid aptamer screening technology services, to help each customer's scientific research.
How to Order?
If you have any questions regarding our services or products, please feel free to contact us by E-mail: [email protected] or Tel: +86-400-621-6806