2024-02-18 Hits(263)

Introduction to Phage Antibody Library Technology Platform
|
KMD Bioscience can use phage vectors to construct recombinant antibody libraries for humans, mice, rabbits, chickens, alpacas, and others, providing customers with efficient antibody screening and production services. In recent years, new recombinant antibodies have been widely used in the treatment, diagnosis, and research fields of diseases. Among them, phage display technology with rich diversity of antibody genes plays an important role. KMD Bioscience has high-quality M13, T4, and T7 phage display technology platforms, and will select suitable phages for display according to customer needs, providing them with the preparation services of recombinant antibodies such as scFv, Fab, VHH, etc.
|
|
The Process of Phage Antibody Library Technology
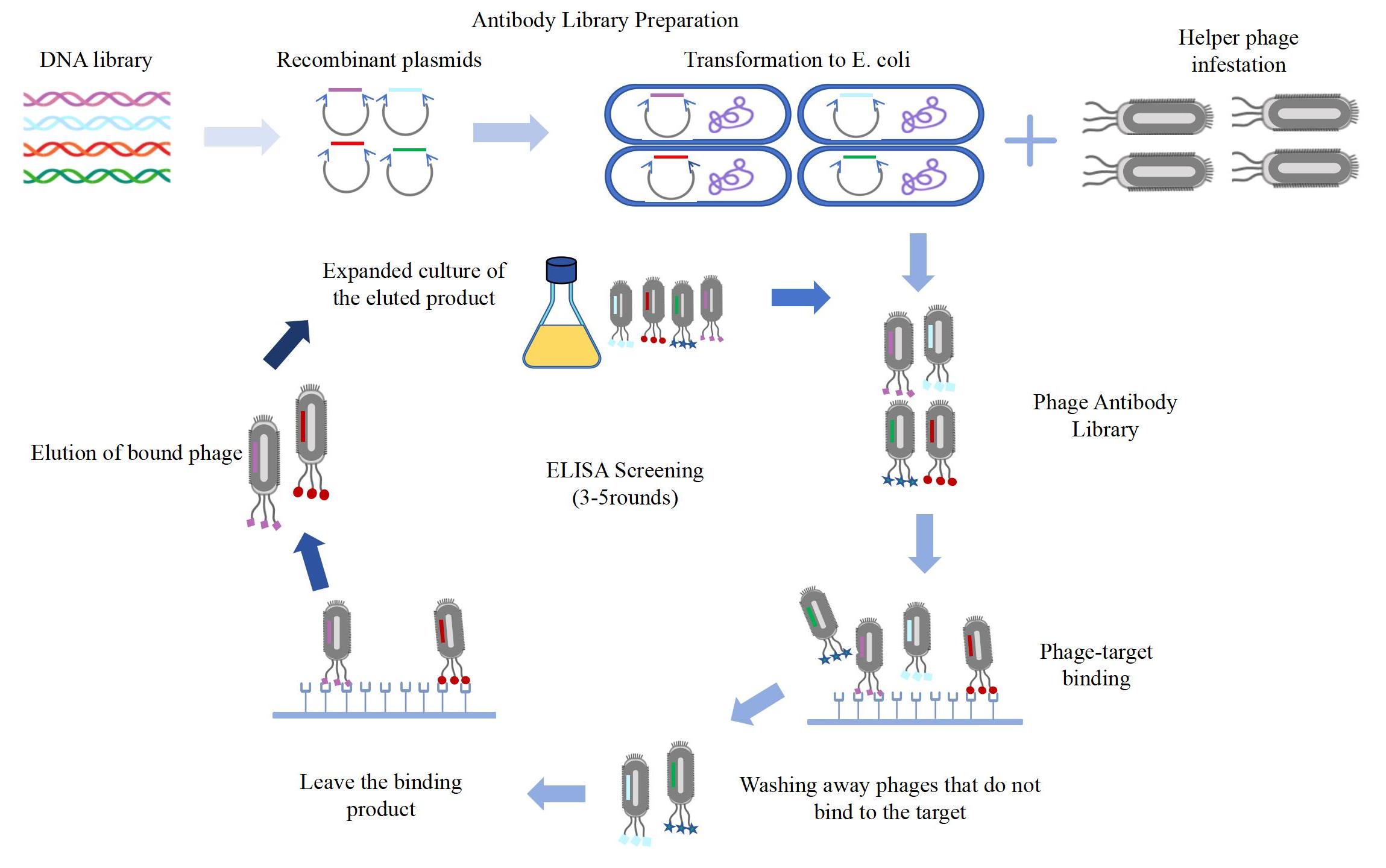
Related Services of Phage Antibody Library Technology
KMD Bioscience has been researching in the field of antibodies for many years. Our phage display technology services cover various library types and can provide customers with multi-species monoclonal antibody production services based on phage-only antibody library platform technology. KMD Bioscience has rich experience in antibody engineering construction and can provide stereoscopic antibody upstream and downstream services.
|
|
Recommended Special Services
KMD Bioscience can provide customers with antibody related services, and its technology platform provides customers with higher quality services.
 |
 |
 |
|
|
|
|
Phage Antibody Library Technology Platforms Related Technical Service Resources
We are willing to share with you the rich technology and resources used in phage antibody library technology platforms technical services to solve your doubts. If you need to view more, please refer to our related articles.
|
|
|
|
|
|
FAQ-Phage Display Technology
Q1: What is phage display?
A: Phage display is a selection technique in which a library of variants of a peptide or protein is expressed on the outside of a phage virion, while the genetic material encoding each variant resides on the inside. This creates a physical linkage between each variant protein sequence and the DNA encoding it, which allows rapid partitioning based on binding affinity to a given target molecule (antibodies, enzymes, cell-surface receptors, etc.) by an in vitro selection process called "biopanning". Biopanning is carried out by incubating the pool of phage-displayed variants with a target of interest that has been immobilized on a plate or bead, washing away unbound phage, and eluting specifically bound phage. The eluted phage is then amplified in vivo and the process repeated, resulting in stepwise enrichment of the phage pool in favor of the tightest binding sequences. After 3-4 rounds of selection/amplification, individual clones are characterized by DNA sequencing.
Q2: What are the advantages of phage display screening over other antibody development method?
A: In comparison with hybridoma technology, phage display technology offers great advantages. Hybridoma method is only available in mouse, rat, hamster and guinea pig. On the other hand, phage display could be used to generate high-affinity monoclonal antibodies from all popular antibody production species, including but not limited to human, mouse, rat, rabbit, chicken, llama, camel, alpaca, cow, dog, sheep, monkey, and shark. Hybridoma-based monoclonal antibody development can only generate a small number of antibody candidates against a particular immunogen at a time; whereas phage display technology can present the entire antibody repertoire (e.g. 10^8) of an immunized animal, in which almost 10% of the antibodies are immunogen(s)-specific. With such a huge pool of potential binders, the chance is much better to use phage display technology to discover antibodies of the desired properties. In addition, using hybridoma technology, it is hard to incorporate an enriching step that can selectively isolate antibodies with the desired functionality. In most cases, all the hybridoma clones are produced first and then validated one by one. In contrast, phage display technology allows various enriching strategies: antibodies of desired properties can be enriched thus those without the desired functionality can be excluded from further validation.
Q3: What research directions can phage display technology be applied to?
A: According to the different display molecules, it can be roughly divided into the following two categories:
1) Protein/antibody: diagnostics, drug navigation, protein structure analysis, humanized modification, and customization of scientific research antibodies.
2) Random peptide library: protein-protein (ligand receptor, information transmission, antagonist/inhibitor, etc.), protein-DNA, diagnostics, neutralization activity, drug and drug navigation, enzyme, and substrate.
Q4: How can the screening process displayed by bacteriophages ensure that the antibodies that are screened for interference can retain the desired antibodies?
A: The principle of selection is actually an experimental process based on the specific reaction between antigens and antibodies. Specifically, the specific binding between the antibody sequence displayed on the surface of the phage and the antigen makes the specific antibody sequence continuously enriched. However, in the process of screening, there must be non-specific binding between bacteriophages of 10^12 or higher and antigens, so after the screening is over, another ELISA identification will be carried out to exclude the non-specific binding antibody clones. Secondly, if there are several highly homologous targets that need to be distinguished at the same time, a closed group can be set up to seal off the cross-binding positive clones in advance, and the remaining phage library can be used to screen specific targets.
Q5: What are the difference among M13, T7, and T4 displaying system?
A: M13 filamentous phage has always been the most popular option and extensively used in various types of research, among many other types of phages. The viral coat consists of five different capsid proteins, including one major capsid pVIII (2,700 copies) and four minor capsids (pIII and pVI at one end while pVII and pIX at the other end). Unlike T4 and T7, M13 is lysogenic phage which is assembled in the periplasm and secreted out of the bacterial membrane without lysing the host. The multiple capsid proteins on M13 phage allow the comprehensive display choices for a variety of peptides and proteins with distinctive characteristics. All five capsids have been successfully used for foreign domain display with unique vectors. pIII and pVIII are mostly the preferred choices for M13 phage display.
Bacteriophage T4 is distinct from M13 in many aspects. T4 has much larger size, tailed structure and double-stranded DNA (dsDNA) genome encoding 50 different proteins. Larger genome DNA allows larger insertions of foreign proteins. Two different domains could be displayed on HOC and SOC, the two non-essential coat proteins. Both N- and C-terminal insertion are available. Without membrane secreting process, host toxicity and confirmation changes could be avoided using T4 phage.
T7 phages have a shorter lifecycle compared to filamentous phages and lambda phages. The assembly of the progeny phages are in bacterial cytoplasm and released by lysis of cell envelope, therefore there is no size limitation and host toxicity resulting from the secreting process. In addition, T7 phages are very stable at extreme conditions where other phages could not survive.